intraflagellar transport 27 homolog (Chlamydomonas)
ZFIN 
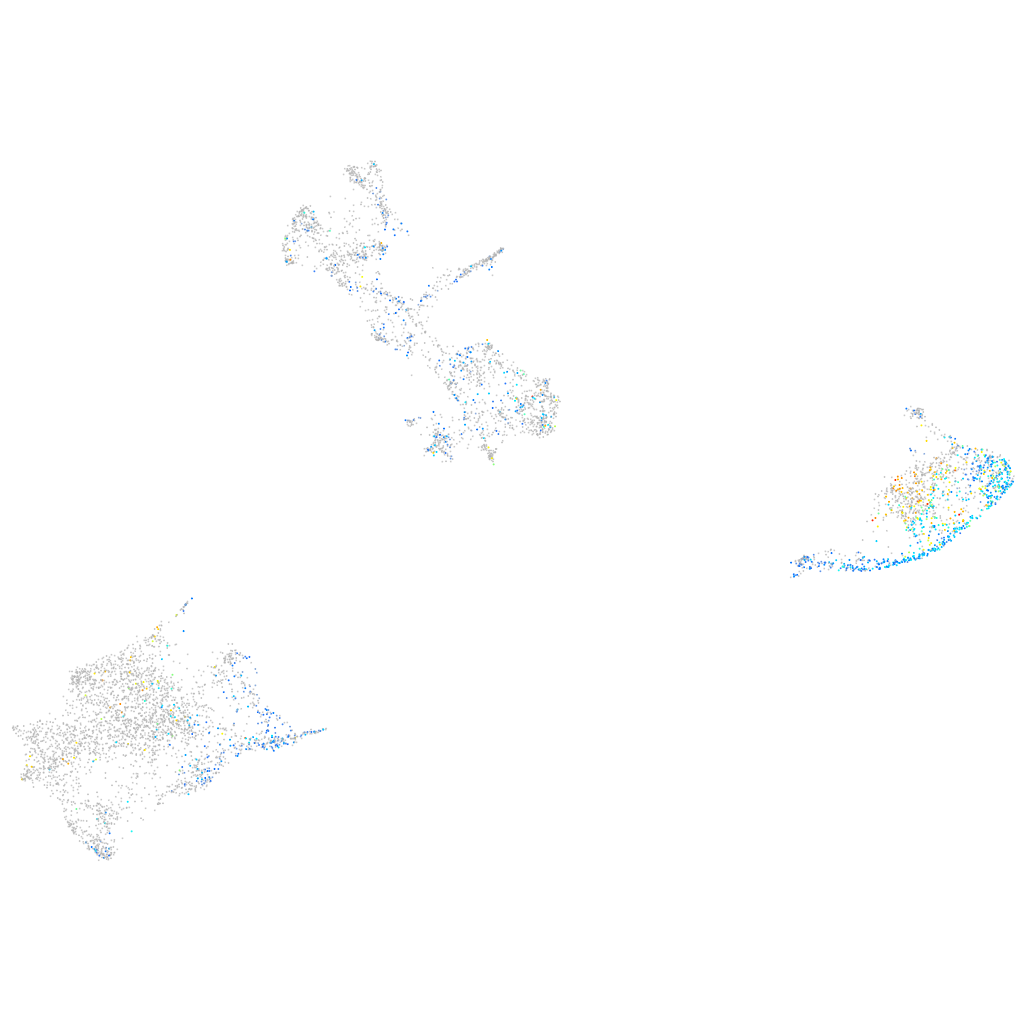
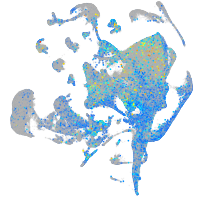
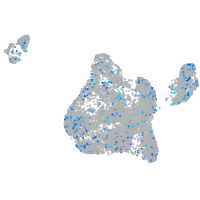
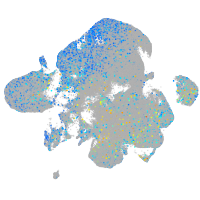
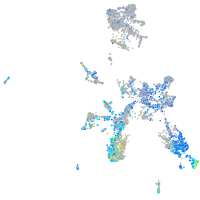
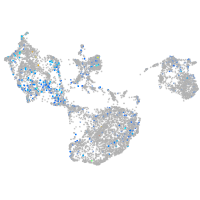
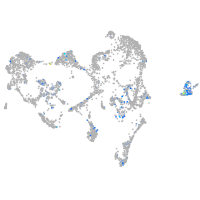
Expression by stage/cluster


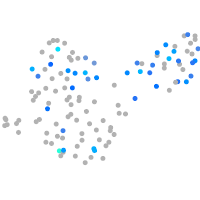
Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dct | 0.451 | CABZ01021592.1 | -0.235 |
| tyrp1a | 0.447 | paics | -0.225 |
| tyrp1b | 0.443 | uraha | -0.213 |
| pmela | 0.439 | si:dkey-251i10.2 | -0.191 |
| zgc:91968 | 0.427 | mdh1aa | -0.181 |
| si:ch73-389b16.1 | 0.407 | phyhd1 | -0.180 |
| oca2 | 0.406 | tmem130 | -0.173 |
| mtbl | 0.386 | slc2a15a | -0.162 |
| slc24a5 | 0.384 | si:ch211-251b21.1 | -0.161 |
| slc45a2 | 0.383 | krt18b | -0.151 |
| kita | 0.358 | glulb | -0.141 |
| aadac | 0.356 | bscl2l | -0.138 |
| prkar1b | 0.356 | pmp22b | -0.133 |
| anxa1a | 0.356 | impdh1b | -0.132 |
| tyr | 0.350 | aox5 | -0.128 |
| gstt1a | 0.337 | aqp3a | -0.128 |
| slc37a2 | 0.320 | atic | -0.127 |
| tspan36 | 0.313 | defbl1 | -0.122 |
| qdpra | 0.312 | pax7b | -0.120 |
| slc3a2a | 0.311 | apoda.1 | -0.118 |
| tspan10 | 0.305 | bco1 | -0.117 |
| slc29a3 | 0.303 | si:ch211-256m1.8 | -0.113 |
| SPAG9 | 0.303 | TMEM19 | -0.113 |
| tmem243b | 0.303 | gpnmb | -0.113 |
| slc22a2 | 0.294 | aldob | -0.112 |
| kcnj13 | 0.293 | cyb5a | -0.112 |
| lamp1a | 0.293 | prps1a | -0.111 |
| tfap2e | 0.288 | lypc | -0.109 |
| atp6v0a2b | 0.281 | unm-sa821 | -0.106 |
| mitfa | 0.280 | sult1st1 | -0.106 |
| pah | 0.278 | oacyl | -0.106 |
| pknox1.2 | 0.276 | prdx5 | -0.105 |
| slc24a4a | 0.275 | si:dkey-197i20.6 | -0.102 |
| slc39a10 | 0.275 | rpz5 | -0.100 |
| atp6v0ca | 0.274 | zgc:113337 | -0.100 |