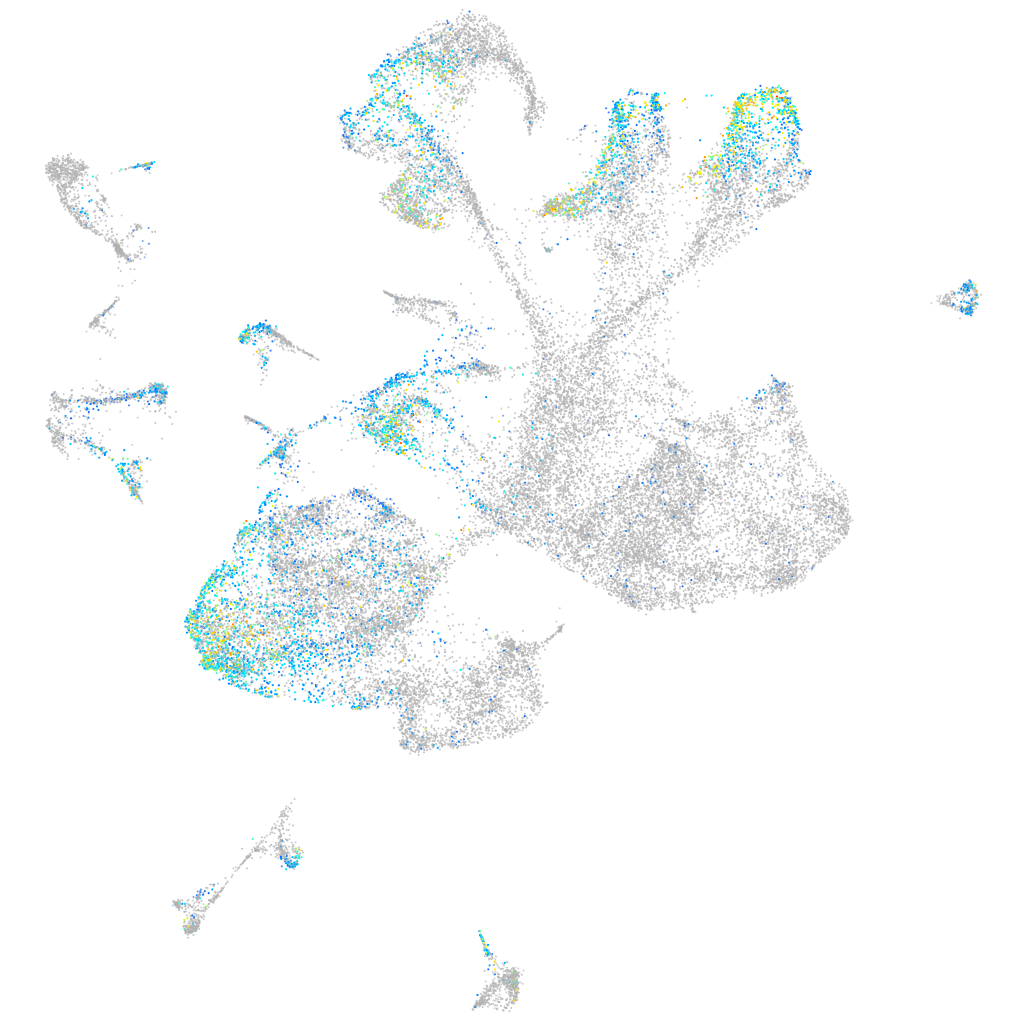
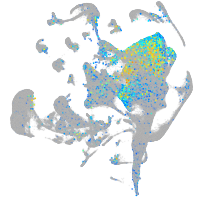
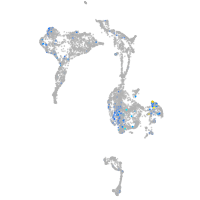
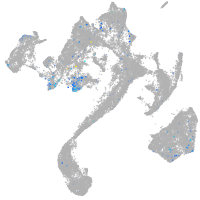
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.379 | rplp2l | -0.349 |
| ndrg3a | 0.373 | si:ch211-222l21.1 | -0.340 |
| aldocb | 0.364 | rplp1 | -0.339 |
| cx43 | 0.322 | rps20 | -0.327 |
| sncgb | 0.308 | hmga1a | -0.327 |
| slc6a1b | 0.308 | si:dkey-151g10.6 | -0.325 |
| ckbb | 0.300 | rps12 | -0.319 |
| si:ch211-66e2.5 | 0.295 | hmgb2b | -0.317 |
| slc1a2b | 0.293 | rps19 | -0.316 |
| slc4a4a | 0.289 | eef1a1l1 | -0.306 |
| pygmb | 0.286 | rpl37 | -0.305 |
| slc6a11b | 0.285 | rpl23 | -0.303 |
| vsnl1b | 0.284 | rps28 | -0.302 |
| nptna | 0.279 | rpl11 | -0.302 |
| snap25a | 0.276 | rps15a | -0.300 |
| tpi1b | 0.275 | rps14 | -0.297 |
| mfge8a | 0.275 | rps21 | -0.296 |
| hepacama | 0.274 | rps9 | -0.294 |
| calm3a | 0.270 | rps7 | -0.294 |
| mgll | 0.268 | rps23 | -0.294 |
| calm1b | 0.266 | rpsa | -0.293 |
| eno2 | 0.266 | rpl36a | -0.293 |
| ndufa4 | 0.265 | rpl27 | -0.292 |
| sypb | 0.265 | rpl26 | -0.291 |
| NPAS3 | 0.264 | rps27a | -0.291 |
| glula | 0.262 | rpl21 | -0.291 |
| slc3a2a | 0.262 | hmgb2a | -0.290 |
| ptn | 0.261 | rpl39 | -0.290 |
| ca4a | 0.259 | rpl8 | -0.290 |
| gpr37l1b | 0.257 | rps27.1 | -0.290 |
| atp6v0cb | 0.255 | rpl12 | -0.290 |
| hspb15 | 0.254 | rpl10a | -0.288 |
| si:ch73-119p20.1 | 0.254 | rps24 | -0.288 |
| cspg5b | 0.253 | rpl35a | -0.288 |
| spock2 | 0.253 | rps25 | -0.284 |