heterogeneous nuclear ribonucleoprotein A0a
ZFIN 
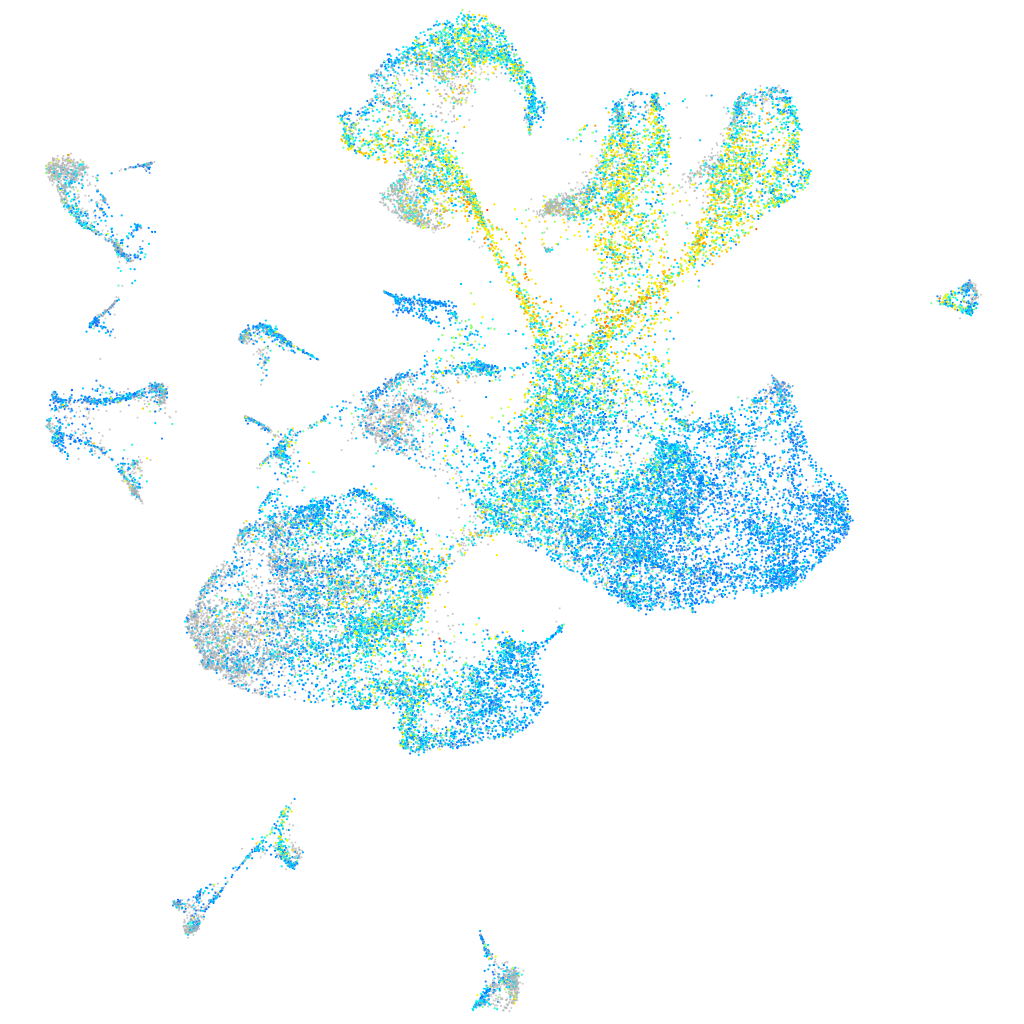
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.424 | atp1a1b | -0.291 |
| cirbpb | 0.385 | glula | -0.270 |
| ptmab | 0.382 | cx43 | -0.257 |
| elavl3 | 0.379 | si:ch211-66e2.5 | -0.238 |
| khdrbs1a | 0.364 | cebpd | -0.233 |
| h3f3d | 0.361 | ptn | -0.229 |
| hnrnpa0l | 0.357 | slc4a4a | -0.228 |
| hmgb3a | 0.356 | mt2 | -0.227 |
| tmeff1b | 0.356 | cd63 | -0.225 |
| tubb2b | 0.355 | ptgdsb.2 | -0.225 |
| myt1b | 0.353 | cox4i2 | -0.224 |
| tubb5 | 0.353 | slc3a2a | -0.223 |
| chd4a | 0.348 | dap1b | -0.222 |
| marcksb | 0.344 | fabp7a | -0.218 |
| hmgn6 | 0.340 | ppap2d | -0.215 |
| hnrnpa0b | 0.338 | efhd1 | -0.215 |
| myt1a | 0.326 | qki2 | -0.213 |
| tuba1c | 0.323 | sparc | -0.213 |
| si:ch211-222l21.1 | 0.319 | apoa2 | -0.202 |
| jagn1a | 0.316 | hepacama | -0.200 |
| si:ch211-288g17.3 | 0.315 | zgc:153704 | -0.199 |
| ilf2 | 0.315 | gpr37l1b | -0.191 |
| nhlh2 | 0.314 | ptgdsb.1 | -0.188 |
| si:dkey-276j7.1 | 0.312 | mfge8a | -0.187 |
| jpt1b | 0.308 | slc1a2b | -0.186 |
| smarce1 | 0.307 | atp1b4 | -0.185 |
| gng2 | 0.303 | apoa1b | -0.184 |
| nova2 | 0.302 | cyp2ad3 | -0.183 |
| hspa8 | 0.301 | cdo1 | -0.182 |
| hnrnpa1a | 0.298 | cyp3c1 | -0.181 |
| cfl1 | 0.296 | sept8b | -0.180 |
| hsp90ab1 | 0.295 | hspb15 | -0.178 |
| tp53inp2 | 0.294 | itm2ba | -0.177 |
| syncrip | 0.291 | id1 | -0.176 |
| nucks1a | 0.291 | acbd7 | -0.173 |