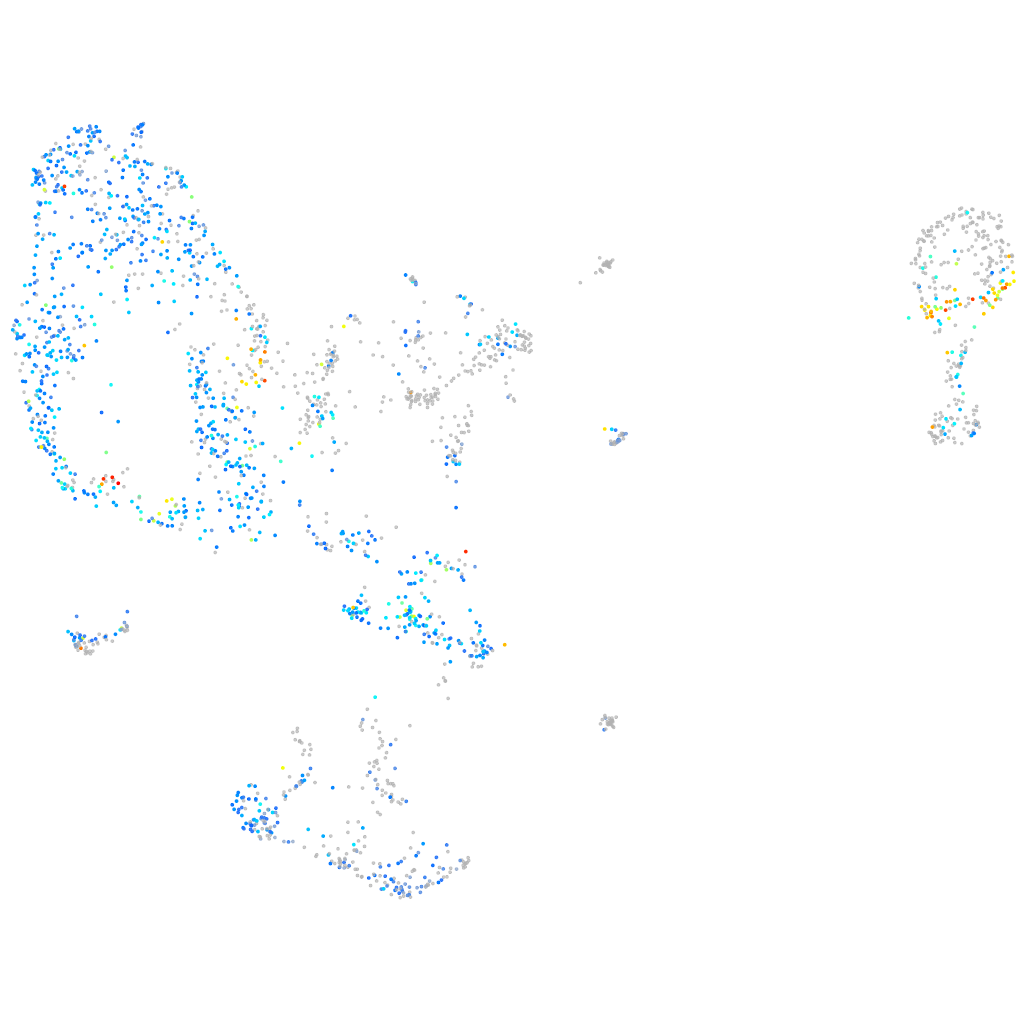
HNF1 homeobox Bb
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| acy3.2 | 0.298 | si:dkey-36i7.3 | -0.184 |
| rbm47 | 0.294 | gapdhs | -0.181 |
| slc26a6 | 0.283 | tmsb1 | -0.170 |
| kcnj13 | 0.273 | si:ch73-359m17.9 | -0.167 |
| hnf1ba | 0.266 | soul2 | -0.159 |
| etnk1 | 0.262 | rnf183 | -0.156 |
| adka | 0.261 | slc12a3 | -0.155 |
| aspa | 0.257 | fabp3 | -0.154 |
| ggt1b | 0.251 | stmn1b | -0.151 |
| ezra | 0.250 | tuba1c | -0.151 |
| slc26a1 | 0.249 | nova2 | -0.148 |
| lrpap1 | 0.240 | tbx2b | -0.148 |
| satb2 | 0.237 | krt4 | -0.146 |
| mfsd4ab | 0.231 | bsnd | -0.145 |
| itga6a | 0.229 | ndrg2 | -0.145 |
| slc5a8l | 0.229 | cdx1a | -0.144 |
| slc4a4a | 0.227 | clcnk | -0.144 |
| st3gal7 | 0.226 | elavl3 | -0.142 |
| si:dkey-194e6.1 | 0.224 | ckbb | -0.142 |
| emx2 | 0.223 | AL954322.2 | -0.142 |
| clic5b | 0.222 | agr2 | -0.141 |
| tmprss2 | 0.221 | marcksl1b | -0.138 |
| pdzk1ip1 | 0.221 | rarga | -0.138 |
| etv5a | 0.218 | rtn1a | -0.138 |
| CABZ01078261.1 | 0.217 | cd63 | -0.138 |
| sptbn5 | 0.216 | mylk5 | -0.137 |
| clptm1 | 0.215 | hbae3 | -0.137 |
| hnf4g | 0.212 | phlda2 | -0.137 |
| ahcyl1 | 0.211 | tmsb | -0.136 |
| ptp4a2b | 0.211 | ahnak | -0.136 |
| dpydb | 0.208 | atp1a1a.3 | -0.135 |
| tpte | 0.208 | si:dkeyp-75h12.5 | -0.134 |
| cubn | 0.208 | trim35-12 | -0.134 |
| pdzk1 | 0.207 | calm1b | -0.134 |
| pcdh1b | 0.207 | gpm6aa | -0.134 |