high mobility group nucleosomal binding domain 3
ZFIN 
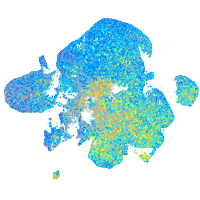
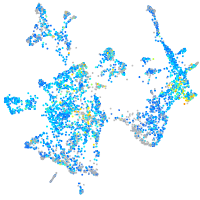
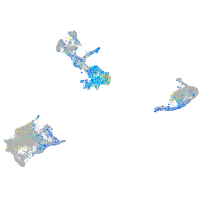
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ssr3 | 0.541 | atp1b1b | -0.373 |
| agr2 | 0.529 | si:dkey-33i11.4 | -0.365 |
| zgc:92380 | 0.517 | COX3 | -0.348 |
| ssr4 | 0.513 | rhbg | -0.344 |
| ssr2 | 0.493 | mt-co1 | -0.338 |
| kdelr2b | 0.489 | mt-co2 | -0.331 |
| nansb | 0.480 | nfe2l2a | -0.324 |
| tspan13a | 0.461 | mt-atp6 | -0.317 |
| foxa3 | 0.454 | mt-nd4 | -0.316 |
| tmed4 | 0.445 | mdh2 | -0.308 |
| pdia3 | 0.445 | mpc1 | -0.307 |
| si:ch73-281n10.2 | 0.432 | abca12 | -0.297 |
| slc39a7 | 0.431 | scel | -0.294 |
| hmgb2a | 0.430 | selenow2b | -0.294 |
| sec11a | 0.427 | si:dkey-247k7.2 | -0.292 |
| pou2f3 | 0.421 | idh2 | -0.290 |
| hmgn2 | 0.421 | mt-nd3 | -0.288 |
| pdia6 | 0.420 | gstp1 | -0.286 |
| ppib | 0.419 | eno3 | -0.286 |
| s100u | 0.416 | wu:fj16a03 | -0.283 |
| serp1 | 0.410 | atp5md | -0.282 |
| tram1 | 0.410 | atp5mc3b | -0.275 |
| mir375-2 | 0.409 | epdl1 | -0.272 |
| zgc:153675 | 0.409 | atp5l | -0.272 |
| tmed2 | 0.405 | mt-nd2 | -0.268 |
| p2rx1 | 0.403 | zgc:111983 | -0.268 |
| anxa5b | 0.401 | rgs5b | -0.267 |
| ddost | 0.399 | plekhf1 | -0.265 |
| krtcap2 | 0.396 | dusp2 | -0.265 |
| pdia5 | 0.395 | pvalb8 | -0.263 |
| kdelr3 | 0.394 | slc25a5 | -0.262 |
| fxyd1 | 0.393 | fthl28 | -0.261 |
| ptmab | 0.392 | mt-nd1 | -0.260 |
| si:ch73-234b20.5 | 0.386 | tob1b | -0.258 |
| gnpnat1 | 0.386 | NDUFB1 | -0.256 |