hypermethylated in cancer 1 like
ZFIN 
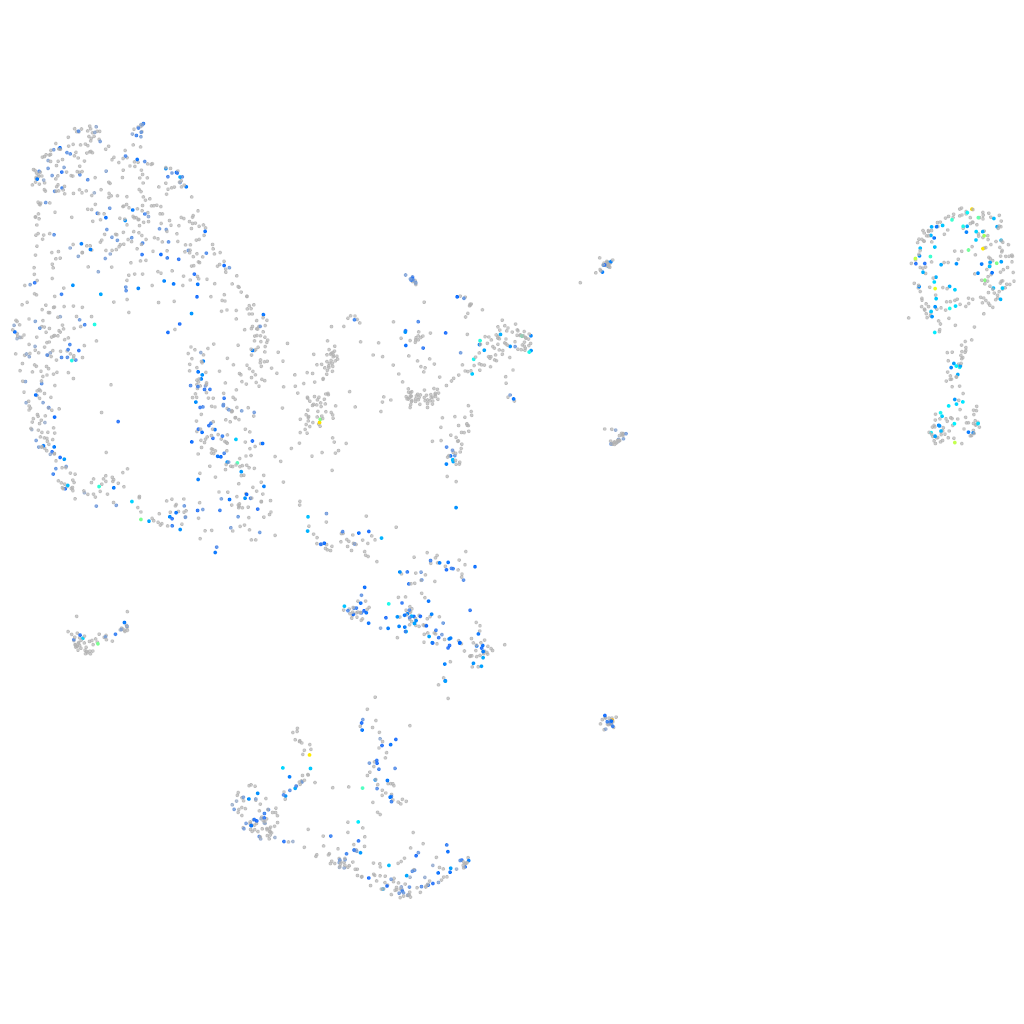
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.189 | rpl37 | -0.157 |
| nid2a | 0.183 | si:ch211-139a5.9 | -0.155 |
| nucks1a | 0.181 | dab2 | -0.144 |
| si:ch211-222l21.1 | 0.173 | aldob | -0.142 |
| hmga1a | 0.172 | rps10 | -0.140 |
| khdrbs1a | 0.171 | gapdh | -0.140 |
| hmgn6 | 0.169 | gstt1a | -0.137 |
| si:ch211-251p5.5 | 0.168 | atp5fa1 | -0.136 |
| ptges3b | 0.167 | gamt | -0.134 |
| eif2s1a | 0.166 | nit2 | -0.133 |
| h2afvb | 0.166 | gstp1 | -0.133 |
| snrpf | 0.165 | grhprb | -0.132 |
| anp32a | 0.163 | zgc:114188 | -0.129 |
| hnrnpl2 | 0.163 | rps17 | -0.128 |
| smc5 | 0.163 | gstr | -0.127 |
| wu:fb97g03 | 0.163 | prdx2 | -0.126 |
| akap12b | 0.161 | fbp1b | -0.126 |
| cirbpa | 0.159 | gcshb | -0.126 |
| hdac1 | 0.158 | zgc:56493 | -0.123 |
| top1l | 0.158 | eno3 | -0.122 |
| smarca4a | 0.158 | atp5mc1 | -0.122 |
| qkia | 0.158 | hoga1 | -0.121 |
| snrpa1 | 0.157 | cox6a2 | -0.121 |
| tspan7 | 0.157 | atp5f1e | -0.121 |
| hmgb2a | 0.157 | gpt2l | -0.120 |
| hnrnpa1a | 0.156 | pnp6 | -0.120 |
| cdx4 | 0.155 | aldh7a1 | -0.117 |
| apoc1 | 0.155 | mdh1aa | -0.116 |
| nr6a1a | 0.154 | atp5f1b | -0.116 |
| prkrip1 | 0.154 | rgn | -0.115 |
| ccnd1 | 0.154 | agxtb | -0.115 |
| CT030188.1 | 0.153 | si:dkey-28n18.9 | -0.115 |
| ilf3b | 0.153 | atp5mc3b | -0.115 |
| hnrnpa1b | 0.153 | sod1 | -0.115 |
| si:ch73-281n10.2 | 0.153 | ldhba | -0.114 |