hypermethylated in cancer 1
ZFIN 
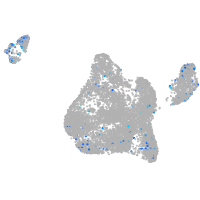
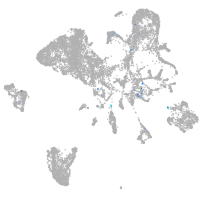
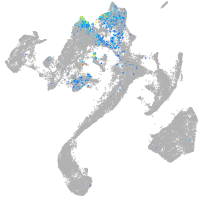
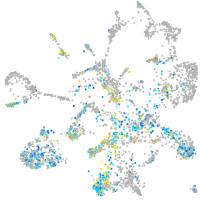
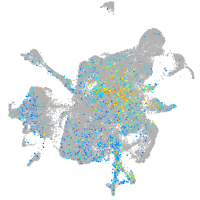
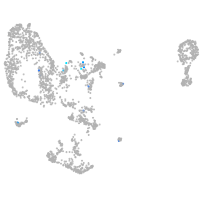
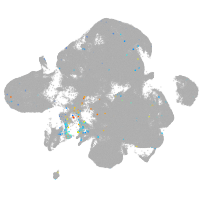
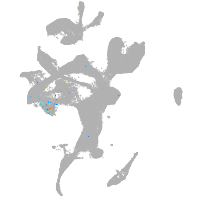
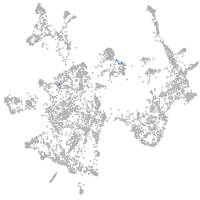
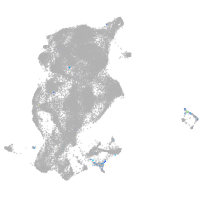
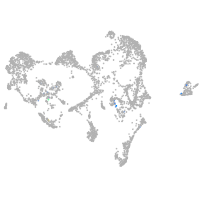
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01074298.1 | 0.163 | rps15a | -0.024 |
| ackr4a | 0.141 | rps3 | -0.022 |
| BX664750.3 | 0.130 | alyref | -0.022 |
| si:ch211-142d6.2 | 0.129 | zgc:114188 | -0.022 |
| zgc:174356 | 0.118 | hsp90ab1 | -0.022 |
| LOC108183930 | 0.115 | cirbpa | -0.021 |
| ripor3 | 0.113 | rplp2l | -0.020 |
| CU914487.1 | 0.106 | tubb2b | -0.020 |
| CR384099.1 | 0.103 | rpl35 | -0.020 |
| mxtx2 | 0.101 | khdrbs1a | -0.019 |
| LOC103911301 | 0.099 | rpl14 | -0.019 |
| CU104782.1 | 0.099 | rpl29 | -0.019 |
| si:ch73-334d15.4 | 0.096 | rps12 | -0.019 |
| CU657976.1 | 0.096 | rps16 | -0.019 |
| col6a2 | 0.095 | akirin1 | -0.018 |
| sycn.3 | 0.094 | cldn5a | -0.018 |
| si:dkey-288a3.2 | 0.093 | rps6 | -0.018 |
| mir214a | 0.092 | rpl28 | -0.018 |
| cd248a | 0.089 | hmgn6 | -0.018 |
| ssuh2.2 | 0.087 | rpl36 | -0.018 |
| larp6b | 0.086 | tuba1a | -0.018 |
| itga2b | 0.083 | cirbpb | -0.017 |
| alx4a | 0.083 | syncrip | -0.017 |
| ifitm1 | 0.082 | rpl38 | -0.017 |
| adgrd1 | 0.082 | map2k6 | -0.017 |
| CABZ01092746.1 | 0.078 | COX3 | -0.016 |
| flnb | 0.078 | rps27.1 | -0.016 |
| LOC110439181 | 0.076 | rpl36a | -0.016 |
| admb | 0.075 | nedd8 | -0.016 |
| mxra8b | 0.075 | eef1a1l1 | -0.016 |
| LOC101885893 | 0.075 | marcksb | -0.016 |
| col6a3 | 0.073 | hnrnpa1b | -0.016 |
| spon2b | 0.072 | kat7b | -0.016 |
| nr2e3 | 0.068 | rps11 | -0.016 |
| gpr1 | 0.067 | rps29 | -0.016 |