"HAUS augmin-like complex, subunit 5"
ZFIN 
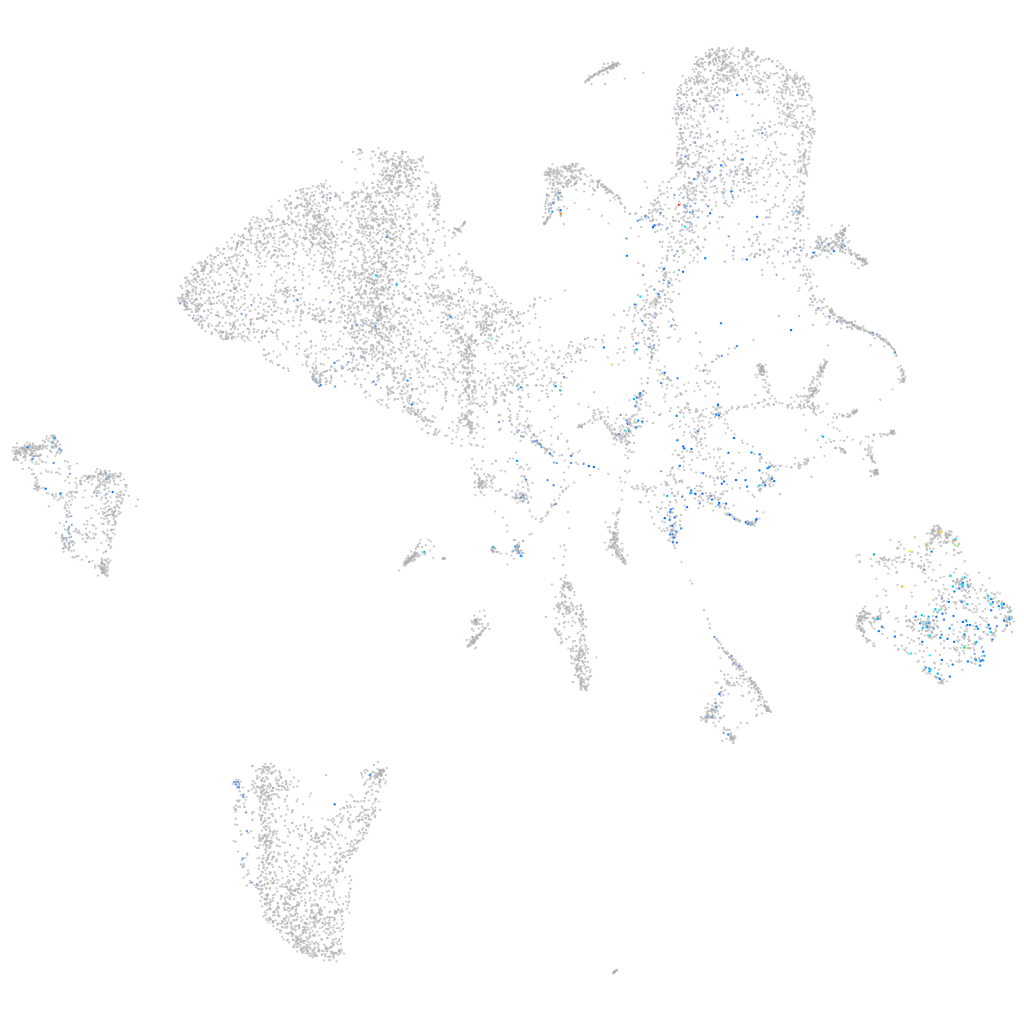
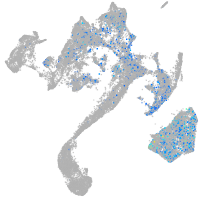
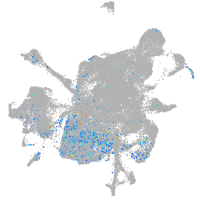
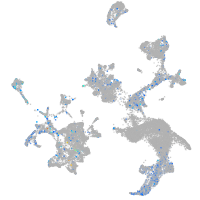
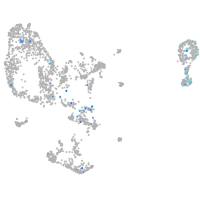
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dek | 0.257 | gamt | -0.143 |
| mki67 | 0.252 | gatm | -0.129 |
| chaf1a | 0.247 | rpl37 | -0.125 |
| lig1 | 0.246 | gapdh | -0.125 |
| stmn1a | 0.243 | zgc:92744 | -0.124 |
| pcna | 0.241 | bhmt | -0.121 |
| brd3a | 0.241 | gpx4a | -0.119 |
| cbx5 | 0.240 | rps10 | -0.119 |
| anp32b | 0.239 | apoa1b | -0.111 |
| ccna2 | 0.239 | ahcy | -0.111 |
| mad2l1 | 0.234 | nme2b.1 | -0.109 |
| sumo3b | 0.234 | zgc:114188 | -0.109 |
| mibp | 0.233 | pnp4b | -0.108 |
| zgc:110425 | 0.231 | apoa4b.1 | -0.108 |
| seta | 0.231 | sod1 | -0.107 |
| fbxo5 | 0.231 | eno3 | -0.107 |
| zgc:110216 | 0.231 | afp4 | -0.106 |
| esco2 | 0.231 | tpi1b | -0.106 |
| hmgb2a | 0.228 | apoa2 | -0.106 |
| hmgb2b | 0.226 | fbp1b | -0.106 |
| hmga1a | 0.226 | pklr | -0.104 |
| rrm1 | 0.225 | abat | -0.104 |
| smc2 | 0.224 | suclg1 | -0.104 |
| nasp | 0.224 | nupr1b | -0.103 |
| banf1 | 0.223 | mat1a | -0.103 |
| ssrp1a | 0.223 | eef1da | -0.102 |
| hnrnpa1b | 0.221 | rps17 | -0.101 |
| CABZ01005379.1 | 0.220 | atp5if1b | -0.101 |
| rbbp4 | 0.219 | agxtb | -0.101 |
| si:ch73-281n10.2 | 0.219 | rbp4 | -0.099 |
| ppm1g | 0.219 | ckba | -0.099 |
| anp32a | 0.218 | dap | -0.099 |
| tubb2b | 0.218 | g6pca.2 | -0.097 |
| tuba8l4 | 0.216 | apobb.1 | -0.097 |
| srsf2a | 0.216 | pgm1 | -0.096 |