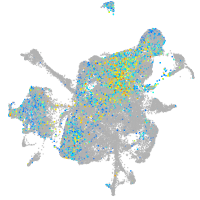
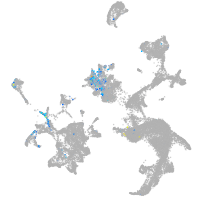
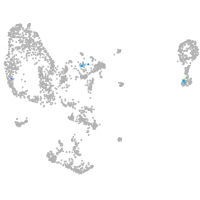
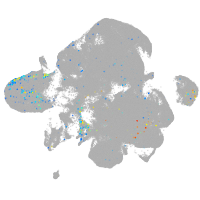
goosecoid
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rnf151 | 0.340 | tspan36 | -0.053 |
| sim2 | 0.280 | rab32a | -0.047 |
| CU929237.1 | 0.267 | smim29 | -0.046 |
| or137-4 | 0.257 | rabl6b | -0.043 |
| si:ch1073-459j12.1 | 0.256 | rnaseka | -0.042 |
| LO018188.1 | 0.255 | slc45a2 | -0.042 |
| prrx1b | 0.247 | atp11a | -0.041 |
| foxf1 | 0.242 | eno3 | -0.041 |
| LOC108179343 | 0.228 | paics | -0.039 |
| barx1 | 0.225 | slc2a15a | -0.039 |
| zgc:153704 | 0.224 | pnp4a | -0.039 |
| foxf2a | 0.221 | XLOC-019062 | -0.038 |
| twist1b | 0.219 | pts | -0.037 |
| CU570682.1 | 0.219 | agtrap | -0.037 |
| ptch1 | 0.217 | abracl | -0.036 |
| wnt5b | 0.215 | tspan10 | -0.036 |
| pdgfra | 0.212 | impdh1b | -0.036 |
| ehd2a | 0.211 | syngr1a | -0.035 |
| si:ch211-145h19.5 | 0.209 | gpr143 | -0.035 |
| prrx1a | 0.209 | actb2 | -0.035 |
| tmc8 | 0.209 | pcbd1 | -0.034 |
| prlhr2a | 0.209 | LOC103910009 | -0.034 |
| ano6 | 0.207 | ldhba | -0.034 |
| col1a2 | 0.202 | mitfa | -0.034 |
| ifitm1 | 0.202 | C12orf75 | -0.034 |
| igf1 | 0.200 | akr1b1 | -0.034 |
| XLOC-022924 | 0.200 | prdx6 | -0.034 |
| c1qb | 0.200 | tpd52 | -0.033 |
| si:dkey-108k21.12 | 0.199 | tmem243a | -0.033 |
| fgfr2 | 0.199 | si:dkey-21a6.5 | -0.033 |
| ecrg4a | 0.198 | tyr | -0.033 |
| CR847534.1 | 0.197 | rab38 | -0.032 |
| sfrp2 | 0.197 | qdpra | -0.032 |
| col5a2a | 0.196 | pik3c3 | -0.032 |
| si:ch211-106h4.12 | 0.192 | stx12 | -0.032 |