galectin-related inter-fiber protein
ZFIN 
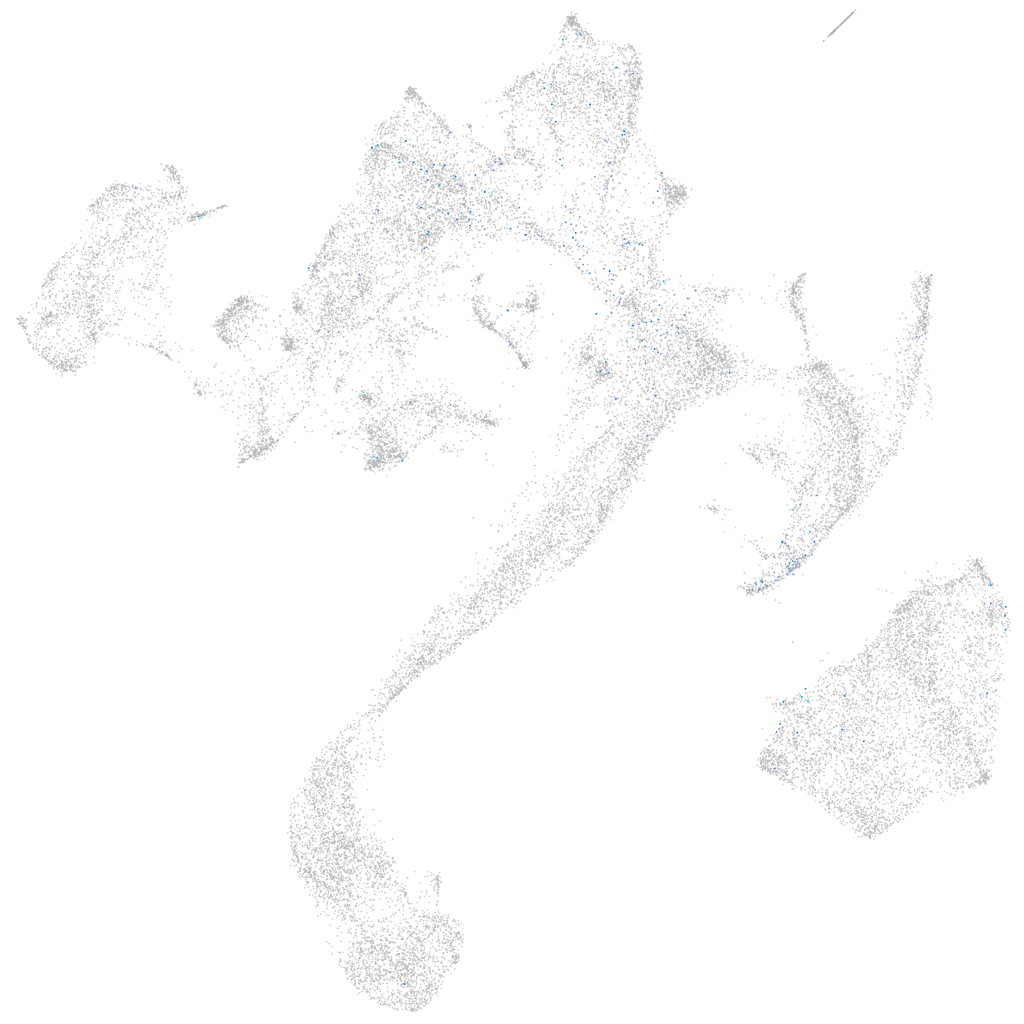
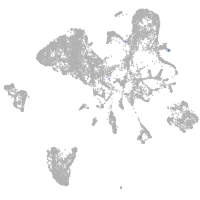
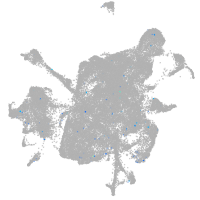
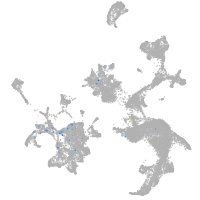
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lrrtm4l2 | 0.110 | aldoab | -0.047 |
| krt93 | 0.105 | cox7c | -0.046 |
| crygm2d14 | 0.103 | atp2a1 | -0.046 |
| hid1b | 0.103 | actc1b | -0.045 |
| sult3st4 | 0.096 | ckma | -0.045 |
| XLOC-036507 | 0.091 | gapdh | -0.045 |
| LOC101885445 | 0.085 | ttn.1 | -0.044 |
| npy2r | 0.084 | nme2b.2 | -0.044 |
| lfng | 0.080 | tmem38a | -0.042 |
| crygm2d5 | 0.080 | pabpc4 | -0.042 |
| hcar1-3 | 0.080 | zgc:101853 | -0.042 |
| LOC103911641 | 0.078 | myl1 | -0.042 |
| taar18j | 0.077 | ckmb | -0.042 |
| BX537288.2 | 0.077 | cox17 | -0.041 |
| LOC108180115 | 0.077 | ttn.2 | -0.041 |
| LOC110439392 | 0.074 | cycsb | -0.041 |
| trim35-25 | 0.073 | tnnt3a | -0.041 |
| wnt16 | 0.070 | srl | -0.040 |
| foxn1 | 0.069 | atp5f1e | -0.040 |
| tpm4a | 0.067 | cox5aa | -0.040 |
| crp7 | 0.066 | gamt | -0.040 |
| mfap2 | 0.066 | mybphb | -0.040 |
| wls | 0.066 | tnnc2 | -0.039 |
| BX901923.2 | 0.066 | neb | -0.039 |
| LOC101884122 | 0.065 | desma | -0.039 |
| itga9 | 0.064 | ak1 | -0.039 |
| si:ch211-156l18.8 | 0.064 | hsp90aa1.1 | -0.039 |
| rhobtb2a | 0.062 | acta1b | -0.039 |
| pax3a | 0.061 | ldb3a | -0.039 |
| marcksl1a | 0.061 | klhl41b | -0.039 |
| nkx3-1 | 0.061 | ldb3b | -0.039 |
| lin28a | 0.061 | actc1a | -0.038 |
| AL954694.2 | 0.061 | si:ch73-367p23.2 | -0.038 |
| mdka | 0.061 | cav3 | -0.038 |
| v2ra18 | 0.060 | atp5md | -0.038 |