"glutamate receptor, ionotropic, AMPA 1a"
ZFIN 
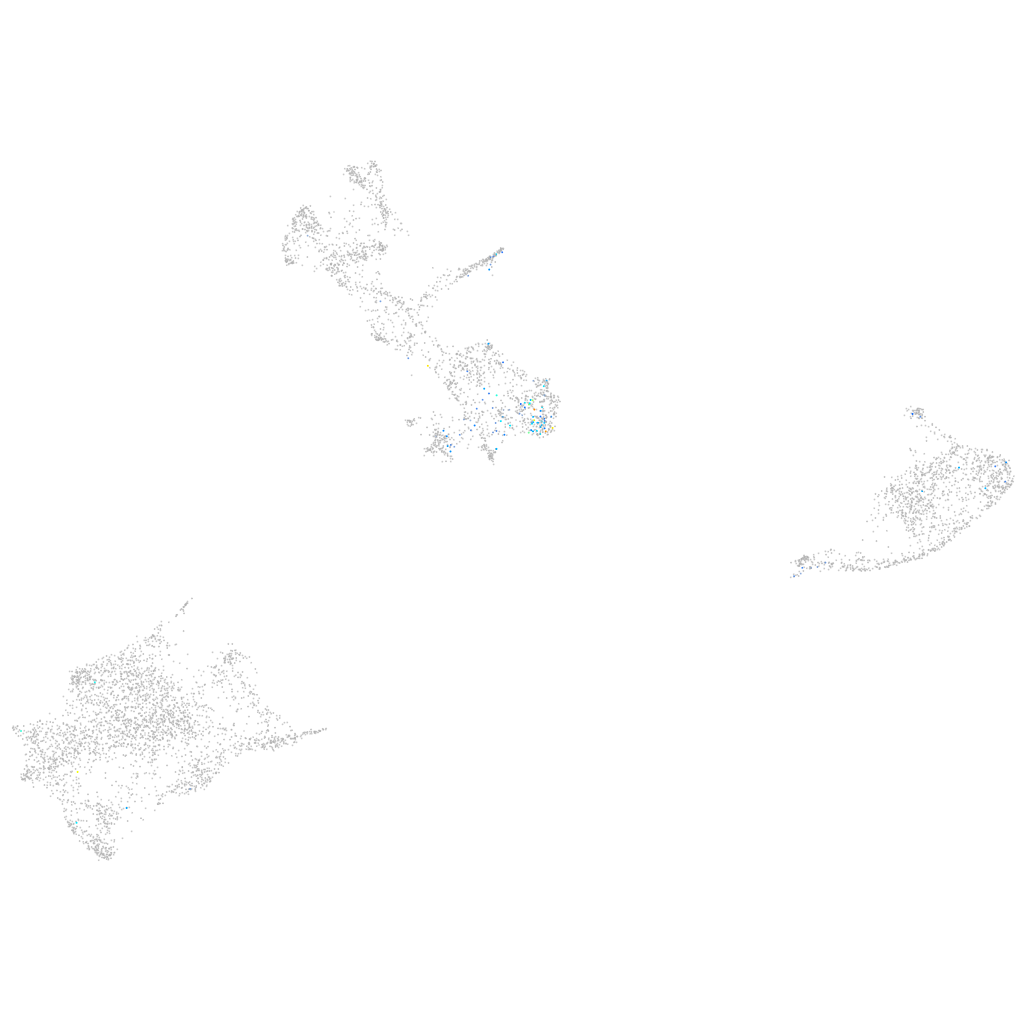
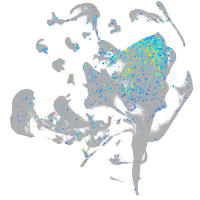
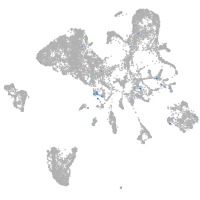
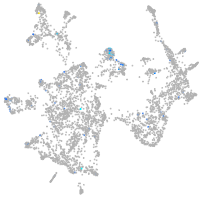
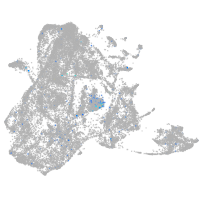
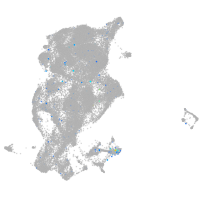
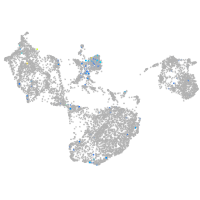
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX649516.5 | 0.258 | tspan36 | -0.066 |
| TMEM200C | 0.245 | paics | -0.064 |
| islr2 | 0.232 | si:dkey-151g10.6 | -0.064 |
| CT027989.1 | 0.230 | eef1a1l1 | -0.064 |
| LOC568754 | 0.220 | zgc:56493 | -0.063 |
| kcnc3a | 0.217 | glulb | -0.061 |
| nrxn2a | 0.216 | prps1a | -0.061 |
| slc6a1a | 0.212 | rps2 | -0.059 |
| syt1a | 0.210 | rplp1 | -0.056 |
| si:ch73-111e15.1 | 0.209 | syngr1a | -0.053 |
| elavl4 | 0.207 | impdh1b | -0.052 |
| cntnap5l | 0.206 | CABZ01032488.1 | -0.051 |
| dicp1.16 | 0.206 | ppiaa | -0.051 |
| chga | 0.204 | rpl11 | -0.050 |
| slc35g2b | 0.202 | rps19 | -0.050 |
| adcy8 | 0.199 | zgc:110239 | -0.049 |
| atp1a3a | 0.194 | uraha | -0.049 |
| atp2b3a | 0.191 | prdx6 | -0.049 |
| slc32a1 | 0.191 | gstp1 | -0.049 |
| nova1 | 0.190 | agtrap | -0.049 |
| CT573793.3 | 0.189 | ctsd | -0.049 |
| cacna2d2b | 0.188 | gpd1b | -0.049 |
| sncb | 0.188 | cyb5a | -0.048 |
| adgrb1a | 0.187 | gmps | -0.048 |
| maptb | 0.185 | rab38 | -0.048 |
| erbb4a | 0.182 | atic | -0.047 |
| xpr1a | 0.180 | actb2 | -0.047 |
| stmn1b | 0.179 | cst14a.2 | -0.047 |
| BX571762.1 | 0.179 | hspa8 | -0.047 |
| CABZ01051901.1 | 0.179 | eno3 | -0.047 |
| nat16 | 0.179 | rps28 | -0.047 |
| stx1b | 0.179 | rab32a | -0.047 |
| glipr1b | 0.179 | gart | -0.046 |
| celf4 | 0.177 | mdh1aa | -0.046 |
| slc6a1b | 0.177 | rplp0 | -0.046 |