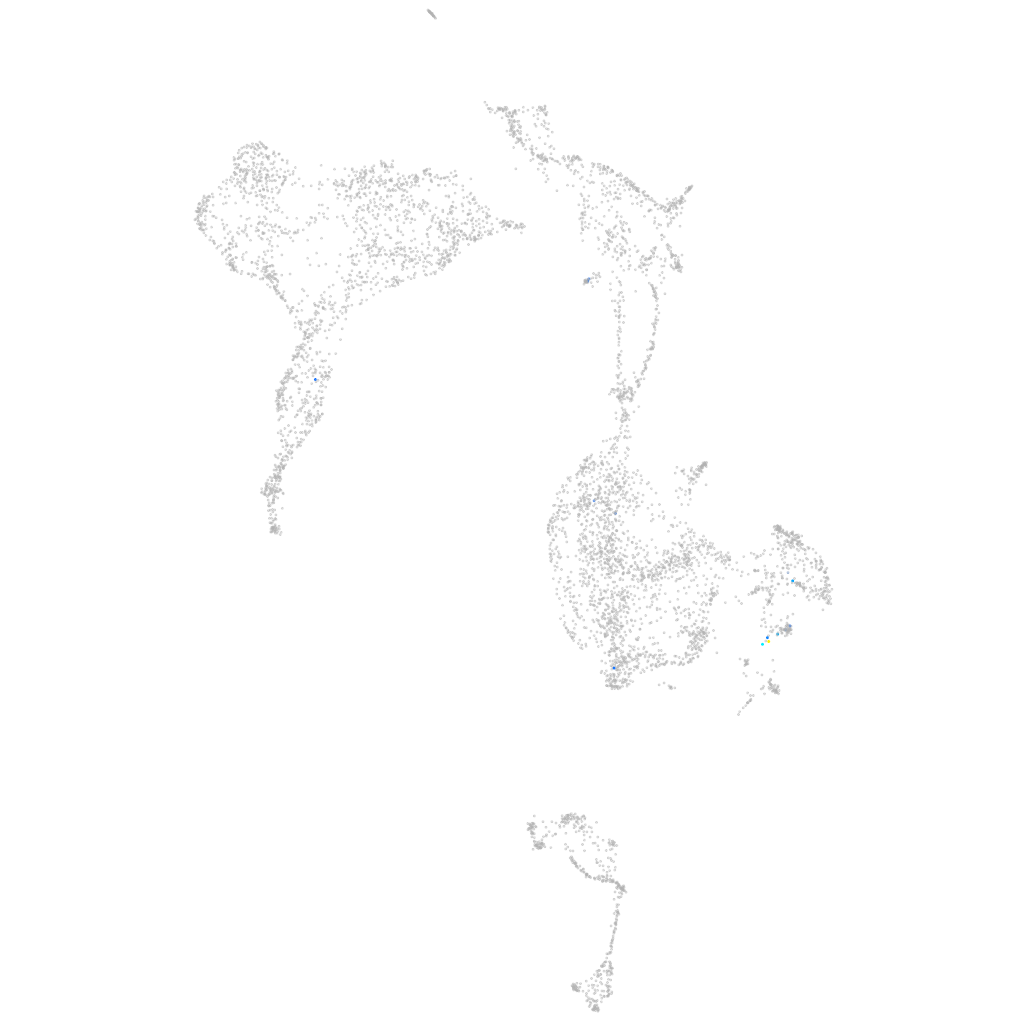
G protein-coupled receptor 182
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mrc1a | 0.771 | sec61b | -0.043 |
| lyve1b | 0.768 | rpl4 | -0.039 |
| ACKR2 | 0.695 | setb | -0.036 |
| myct1a | 0.671 | asph | -0.035 |
| sele | 0.661 | NC-002333.4 | -0.034 |
| si:ch211-145b13.6 | 0.655 | pabpc1a | -0.034 |
| ushbp1 | 0.640 | tmed2 | -0.034 |
| si:dkey-28n18.9 | 0.617 | hspb1 | -0.032 |
| fli1b | 0.616 | rab5if | -0.032 |
| LOC108182616 | 0.616 | odc1 | -0.032 |
| si:dkey-151m22.8 | 0.616 | fth1a | -0.032 |
| plvapb | 0.615 | lrrc59 | -0.031 |
| si:ch211-66e2.3 | 0.604 | abcf1 | -0.031 |
| ecscr | 0.603 | hmga1a | -0.031 |
| si:ch211-202m22.1 | 0.582 | cd2bp2 | -0.030 |
| si:dkeyp-33c10.7 | 0.582 | xbp1 | -0.030 |
| CU463231.2 | 0.582 | hnrnpa0a | -0.030 |
| lyve1a | 0.582 | ufm1 | -0.028 |
| pcdh12 | 0.559 | rpf1 | -0.028 |
| clec14a | 0.551 | fcf1 | -0.027 |
| pecam1 | 0.515 | ssr1 | -0.027 |
| mfng | 0.483 | ssr2 | -0.027 |
| CU927890.1 | 0.462 | dnajc1 | -0.027 |
| shank3b | 0.456 | rad23b | -0.027 |
| etv2 | 0.448 | kdelr2a | -0.027 |
| zgc:153311 | 0.438 | si:ch73-281n10.2 | -0.027 |
| kdr | 0.437 | nop58 | -0.026 |
| cdh5 | 0.429 | sec61a1l | -0.026 |
| exoc3l2a | 0.411 | aldoaa | -0.026 |
| znf521 | 0.410 | srp19 | -0.026 |
| ccm2l | 0.406 | arf5 | -0.026 |
| thsd1 | 0.406 | fkbp11 | -0.026 |
| LOC108180727 | 0.402 | mad2l1 | -0.026 |
| XLOC-039757 | 0.400 | cstf1 | -0.025 |
| entpd8 | 0.398 | hnrnpa0b | -0.025 |