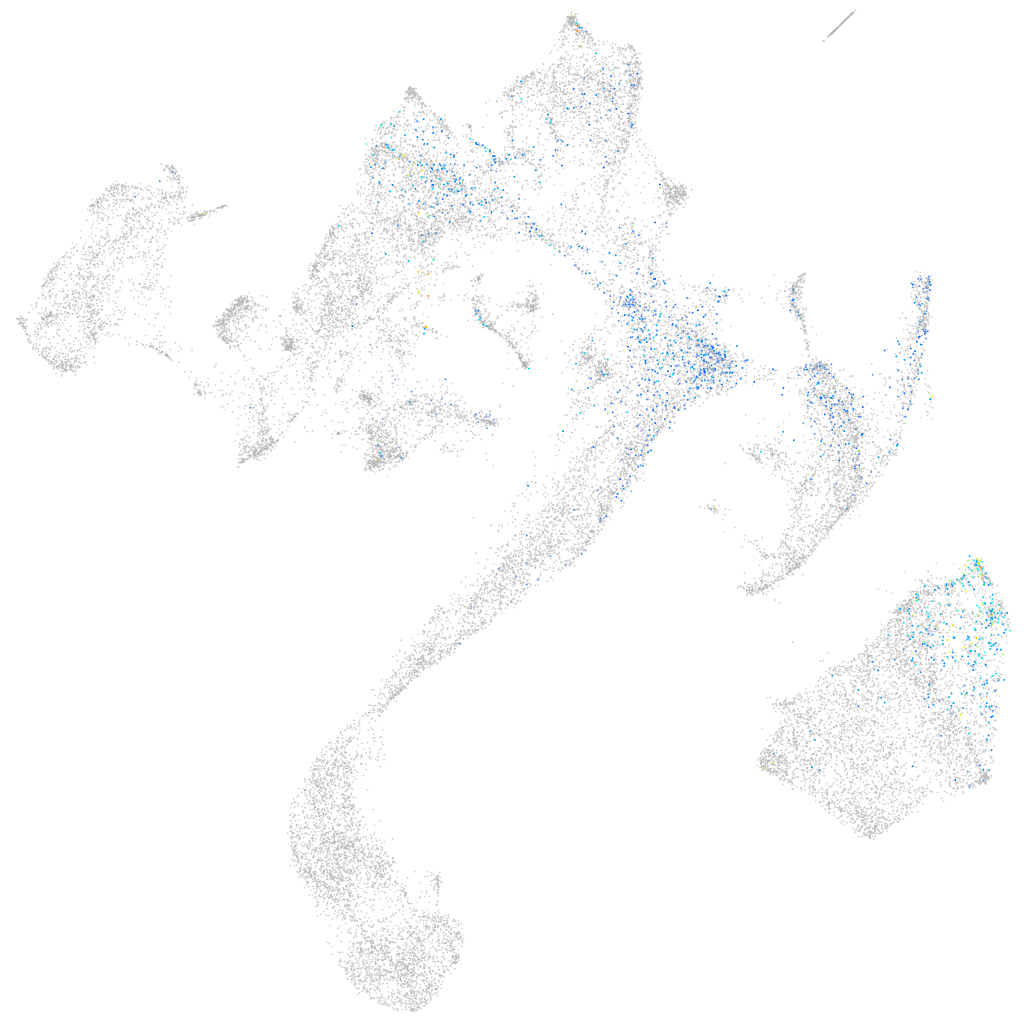
G protein-coupled receptor 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm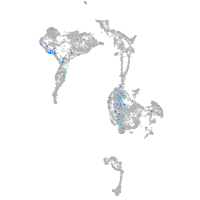 |
muscle 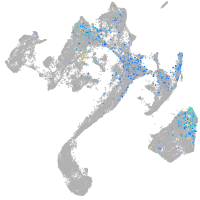 |
mural cells / non-skeletal muscle 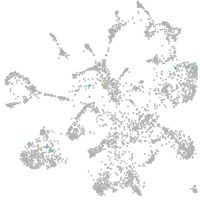 |
mesenchyme 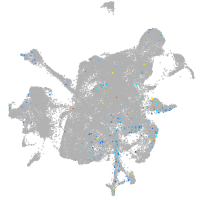 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| asph | 0.211 | ckmb | -0.115 |
| meox1 | 0.172 | actc1b | -0.115 |
| ripply1 | 0.169 | atp2a1 | -0.113 |
| aldh1a2 | 0.162 | ckma | -0.112 |
| rdh10a | 0.159 | tmem38a | -0.109 |
| tcf15 | 0.153 | pabpc4 | -0.108 |
| fn1b | 0.148 | ak1 | -0.107 |
| tbx6 | 0.147 | aldoab | -0.107 |
| dmrt2a | 0.142 | gapdh | -0.105 |
| sfrp1a | 0.142 | mylpfa | -0.102 |
| lpar1 | 0.138 | tnnc2 | -0.100 |
| pcdh10b | 0.137 | ldb3b | -0.099 |
| kazald2 | 0.134 | neb | -0.099 |
| cpn1 | 0.130 | eno3 | -0.099 |
| plp2 | 0.127 | myl1 | -0.098 |
| XLOC-042229 | 0.124 | eno1a | -0.098 |
| CT030188.1 | 0.121 | tnnt3a | -0.097 |
| cdh2 | 0.121 | srl | -0.096 |
| arl4aa | 0.119 | mylz3 | -0.096 |
| si:ch73-281n10.2 | 0.119 | ank1a | -0.096 |
| myf5 | 0.115 | acta1b | -0.095 |
| mdka | 0.115 | cav3 | -0.095 |
| CABZ01075068.1 | 0.114 | CABZ01078594.1 | -0.095 |
| pstpip2 | 0.113 | si:ch73-367p23.2 | -0.094 |
| tjp2b | 0.112 | cox17 | -0.093 |
| XLOC-004280 | 0.111 | nme2b.2 | -0.093 |
| XLOC-040516 | 0.110 | actn3a | -0.093 |
| meis1b | 0.108 | atp1a2a | -0.093 |
| qkia | 0.106 | gamt | -0.093 |
| fkbp7 | 0.106 | myom1a | -0.092 |
| efnb2a | 0.106 | vdac3 | -0.092 |
| BX927258.1 | 0.105 | pgam2 | -0.092 |
| angptl7 | 0.105 | si:ch211-266g18.10 | -0.091 |
| ston2 | 0.104 | mdh2 | -0.091 |
| nectin1b | 0.104 | hhatla | -0.091 |