"glutamic-oxaloacetic transaminase 2b, mitochondrial"
ZFIN 
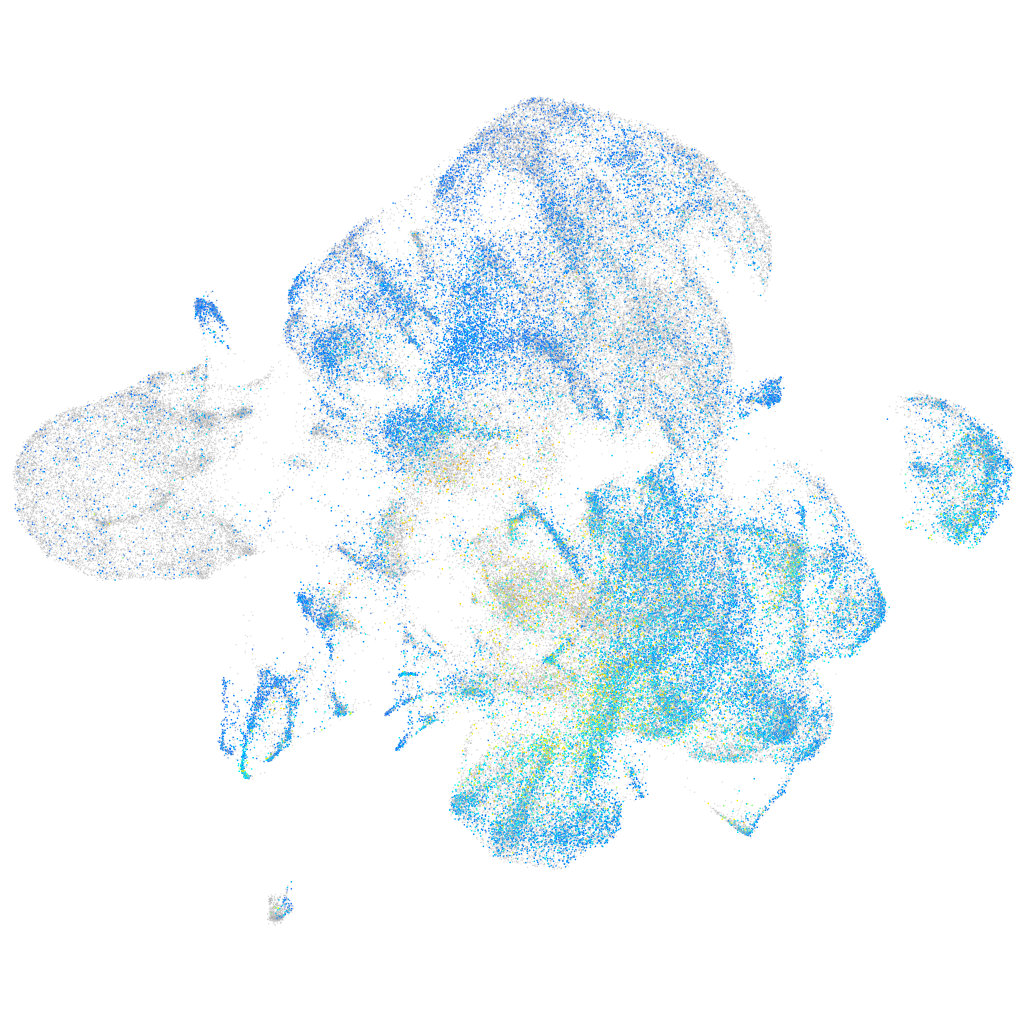
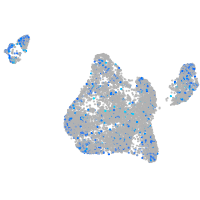
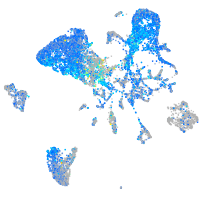
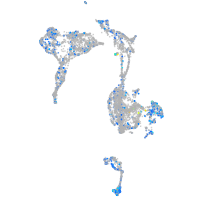
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.272 | hmgb2a | -0.185 |
| tpi1b | 0.255 | hmga1a | -0.162 |
| sncb | 0.246 | hspb1 | -0.154 |
| gng3 | 0.245 | id1 | -0.146 |
| ywhag2 | 0.244 | akap12b | -0.140 |
| atp6v1e1b | 0.241 | sox3 | -0.139 |
| atp6v1g1 | 0.238 | sox19a | -0.133 |
| ckbb | 0.235 | si:ch211-152c2.3 | -0.130 |
| atp5mc1 | 0.235 | sb:cb81 | -0.130 |
| stmn2a | 0.235 | msna | -0.129 |
| ldhba | 0.234 | XLOC-003689 | -0.124 |
| zgc:65894 | 0.232 | XLOC-003690 | -0.122 |
| atp6v0cb | 0.230 | cldn5a | -0.122 |
| calm1a | 0.228 | cx43.4 | -0.121 |
| tuba1c | 0.225 | tspan7 | -0.119 |
| vamp2 | 0.222 | dla | -0.119 |
| stxbp1a | 0.221 | hmgb2b | -0.119 |
| snap25a | 0.220 | si:dkey-42i9.4 | -0.118 |
| calm2a | 0.220 | stm | -0.118 |
| stx1b | 0.218 | apoeb | -0.118 |
| zgc:153426 | 0.217 | lig1 | -0.117 |
| elavl4 | 0.216 | lfng | -0.117 |
| eno1a | 0.216 | notch3 | -0.117 |
| ndufa4 | 0.214 | zgc:110425 | -0.115 |
| tmsb2 | 0.214 | si:dkey-151g10.6 | -0.114 |
| ywhah | 0.209 | NC-002333.4 | -0.114 |
| rtn1b | 0.208 | mki67 | -0.113 |
| ppiab | 0.208 | XLOC-003692 | -0.111 |
| mllt11 | 0.207 | eef1a1l1 | -0.111 |
| cox7a2a | 0.207 | ccnd1 | -0.111 |
| rnasekb | 0.206 | chaf1a | -0.109 |
| cnrip1a | 0.203 | si:ch73-281n10.2 | -0.109 |
| map1aa | 0.202 | apela | -0.109 |
| calm1b | 0.201 | aldob | -0.109 |
| eno2 | 0.201 | nr6a1a | -0.108 |