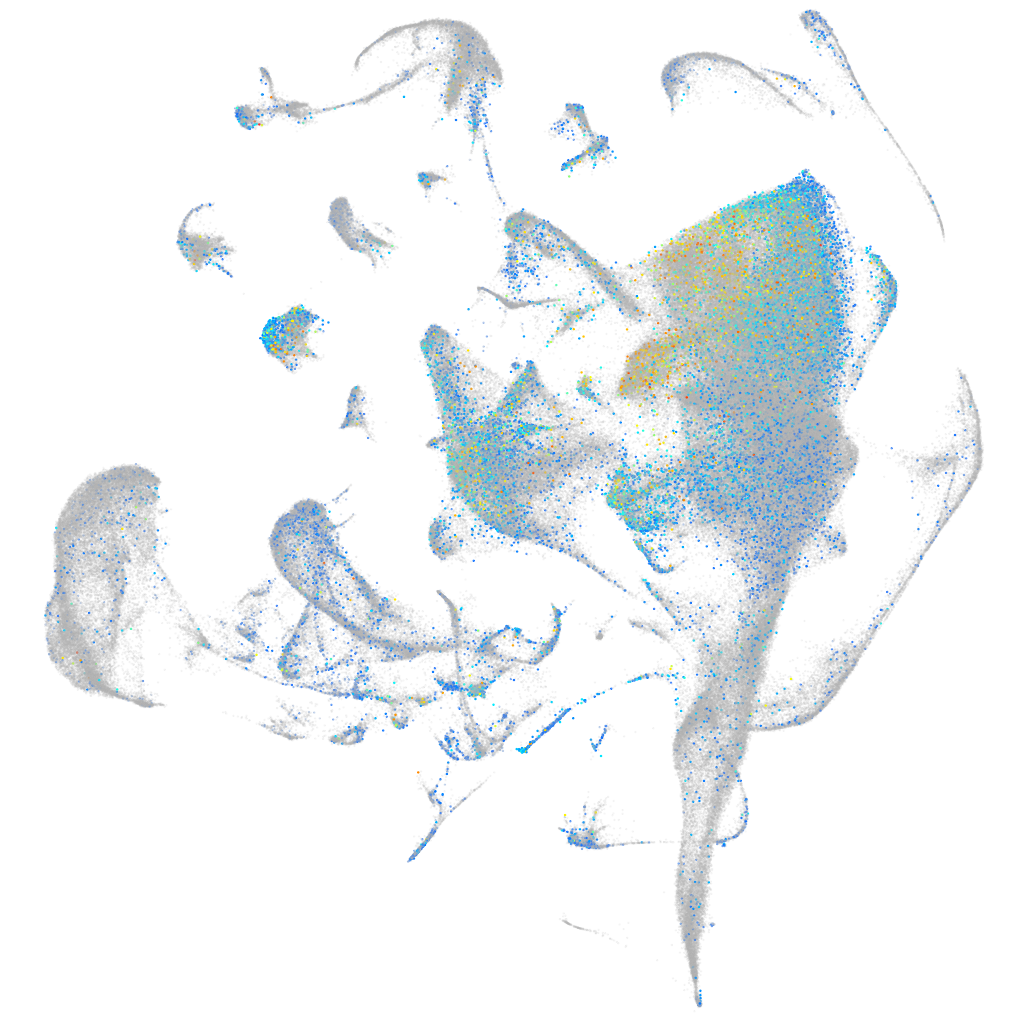glucosamine (N-acetyl)-6-sulfatase a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rnasekb | 0.087 | dkc1 | -0.059 |
| gabarapb | 0.085 | hspb1 | -0.058 |
| rtn1a | 0.084 | nop58 | -0.058 |
| marcksl1a | 0.083 | npm1a | -0.057 |
| si:dkeyp-75h12.5 | 0.082 | fbl | -0.055 |
| gpm6ab | 0.080 | banf1 | -0.054 |
| appb | 0.079 | aldob | -0.053 |
| gng3 | 0.078 | mki67 | -0.053 |
| gpm6aa | 0.078 | pou5f3 | -0.052 |
| calm1a | 0.078 | anp32b | -0.051 |
| atp6v1e1b | 0.077 | lig1 | -0.051 |
| tuba1c | 0.077 | polr3gla | -0.051 |
| vdac3 | 0.077 | ccnb1 | -0.049 |
| atp6v0cb | 0.076 | ddx18 | -0.049 |
| jpt1b | 0.076 | gar1 | -0.049 |
| stmn1b | 0.076 | nop56 | -0.049 |
| tmeff1b | 0.076 | si:dkey-66i24.9 | -0.049 |
| dpysl2b | 0.076 | snu13b | -0.049 |
| elavl3 | 0.075 | tpx2 | -0.049 |
| fam168a | 0.075 | chaf1a | -0.048 |
| rtn1b | 0.075 | lye | -0.048 |
| gapdhs | 0.073 | s100a1 | -0.048 |
| gng2 | 0.073 | zgc:56699 | -0.048 |
| nova2 | 0.073 | bms1 | -0.046 |
| sncb | 0.073 | ccna2 | -0.046 |
| vamp2 | 0.073 | dlgap5 | -0.046 |
| ckbb | 0.072 | nop2 | -0.046 |
| elavl4 | 0.072 | stm | -0.046 |
| fez1 | 0.072 | ube2c | -0.046 |
| stx1b | 0.072 | vox | -0.046 |
| gdi1 | 0.071 | asb11 | -0.045 |
| nsg2 | 0.071 | cdca8 | -0.045 |
| ywhag2 | 0.071 | gnl3 | -0.045 |
| zgc:65894 | 0.071 | ing5b | -0.045 |
| atp6ap2 | 0.070 | nop10 | -0.045 |