guanine monophosphate synthase
ZFIN 
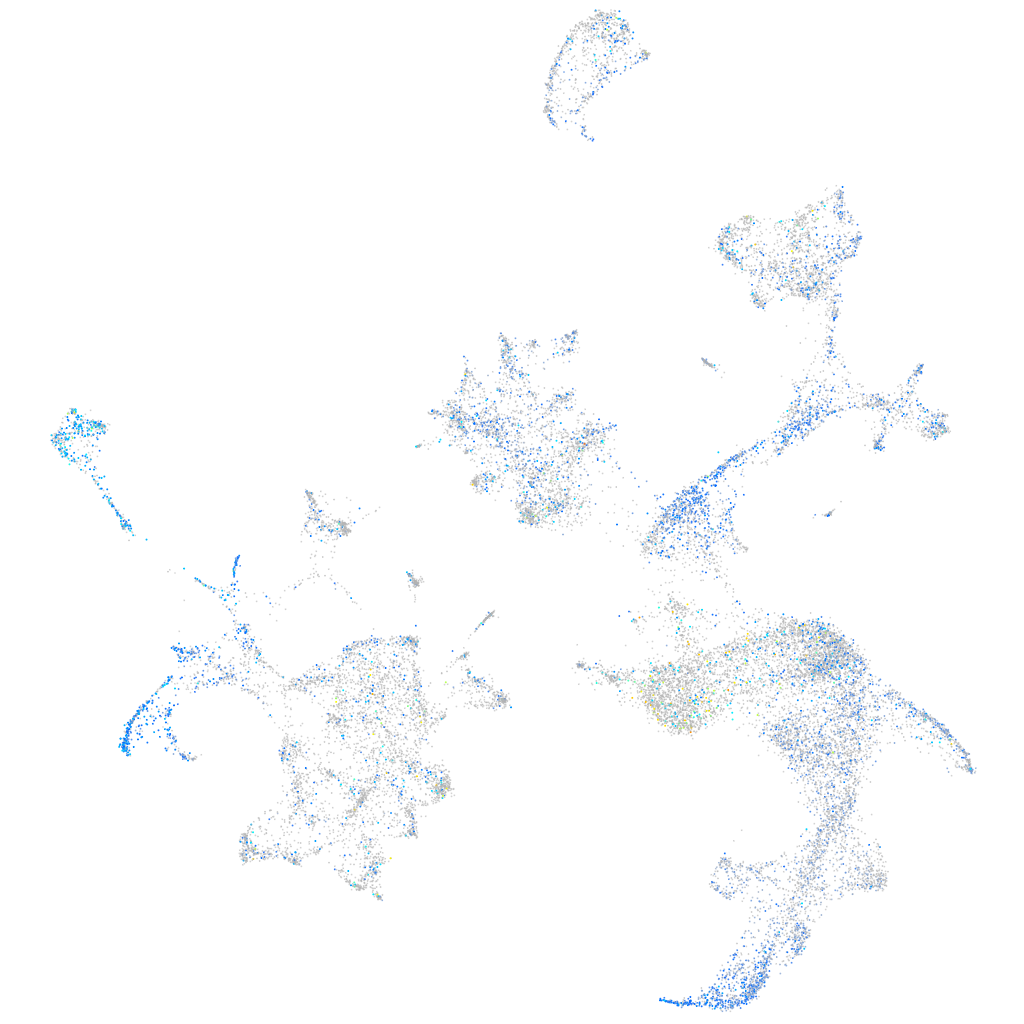
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nop58 | 0.258 | pvalb2 | -0.084 |
| npm1a | 0.257 | krt8 | -0.079 |
| fbl | 0.256 | krt18a.1 | -0.079 |
| nop2 | 0.252 | pvalb1 | -0.078 |
| dkc1 | 0.251 | cpn1 | -0.077 |
| ncl | 0.240 | cdh5 | -0.076 |
| snu13b | 0.239 | cavin2b | -0.073 |
| gnl3 | 0.234 | plvapb | -0.072 |
| nop56 | 0.234 | igfbp7 | -0.072 |
| pprc1 | 0.227 | clec14a | -0.072 |
| nop16 | 0.227 | cldn5b | -0.071 |
| rrp1 | 0.223 | actc1b | -0.071 |
| wdr43 | 0.223 | myct1a | -0.068 |
| bms1 | 0.222 | tie1 | -0.068 |
| ebna1bp2 | 0.220 | kdrl | -0.068 |
| lyar | 0.219 | si:ch211-156j16.1 | -0.067 |
| pes | 0.216 | si:ch211-145b13.6 | -0.067 |
| ptges3b | 0.215 | XLOC-014079 | -0.067 |
| mrto4 | 0.215 | marcksl1a | -0.066 |
| ssb | 0.214 | ramp2 | -0.065 |
| ddx27 | 0.214 | dap1b | -0.065 |
| ppan | 0.213 | ecscr | -0.065 |
| nip7 | 0.212 | srgn | -0.064 |
| rrs1 | 0.210 | pecam1 | -0.063 |
| nolc1 | 0.210 | she | -0.063 |
| llph | 0.209 | plk2b | -0.061 |
| rpl7l1 | 0.208 | tgfbr2b | -0.061 |
| mybbp1a | 0.208 | tspan4b | -0.061 |
| cnbpa | 0.208 | fabp11a | -0.061 |
| si:ch211-217k17.7 | 0.207 | sox18 | -0.061 |
| twistnb | 0.206 | tmsb4x | -0.060 |
| nhp2 | 0.206 | myl9a | -0.060 |
| polr1d | 0.205 | si:dkeyp-97a10.2 | -0.060 |
| nop10 | 0.205 | cav1 | -0.059 |
| gar1 | 0.204 | tagln2 | -0.059 |