"glomulin, FKBP associated protein b"
ZFIN 
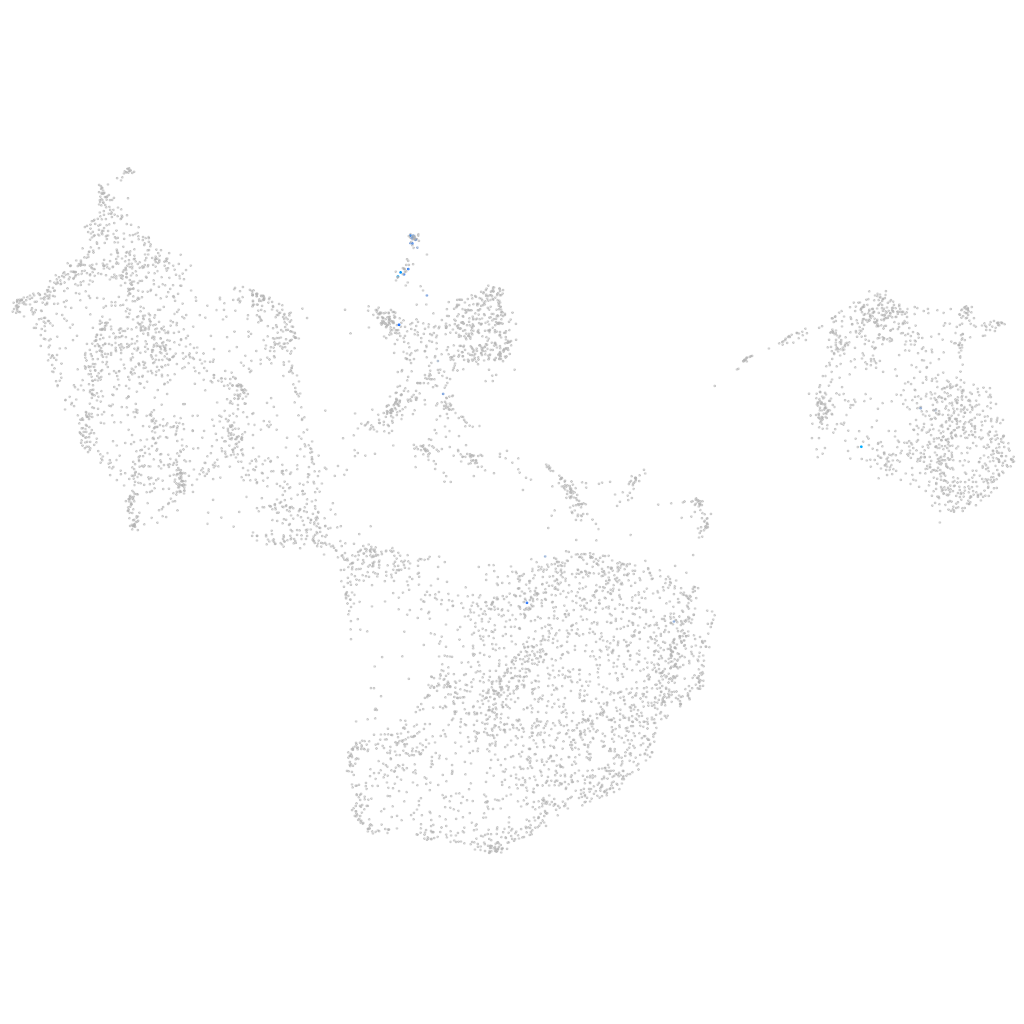
Other cell groups


all cells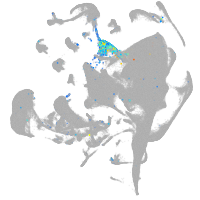 |
blastomeres |
PGCs |
endoderm |
axial mesoderm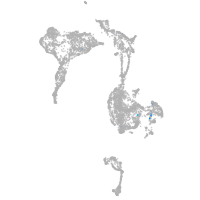 |
muscle  |
mural cells / non-skeletal muscle 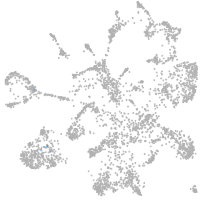 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line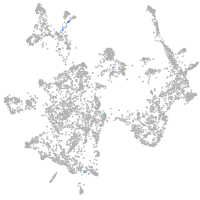 |
epidermis |
periderm |
fin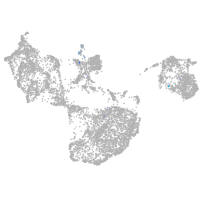 |
ionocytes / mucous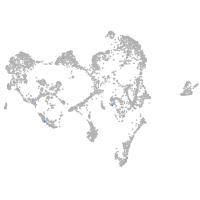 |
pigment cells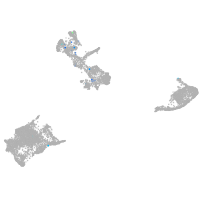 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| abca4b | 0.340 | ctnnd1 | -0.025 |
| cnga3b | 0.302 | pvalb1 | -0.024 |
| chmp6a | 0.298 | pvalb2 | -0.024 |
| si:dkey-283b15.2 | 0.293 | mid1ip1a | -0.023 |
| zgc:112294 | 0.287 | tfap2a | -0.022 |
| elovl4b | 0.280 | si:ch211-166a6.5 | -0.022 |
| mala | 0.266 | txn | -0.022 |
| zgc:103625 | 0.257 | wu:fi04e12 | -0.021 |
| rp1l1a | 0.256 | zgc:174938 | -0.021 |
| si:ch73-28h20.1 | 0.253 | prdx1 | -0.021 |
| mcf2b | 0.244 | si:ch73-238c9.1 | -0.021 |
| si:dkey-220f10.4 | 0.243 | hoxc13a | -0.021 |
| LOC101886519 | 0.240 | tram2 | -0.020 |
| ckmt2a | 0.237 | perp | -0.020 |
| ACVR1C | 0.233 | txndc5 | -0.020 |
| INSM2 | 0.232 | scel | -0.019 |
| pdzph1 | 0.232 | rab13 | -0.019 |
| cnga3a | 0.228 | tead3b | -0.019 |
| paqr8 | 0.228 | ubap2b | -0.019 |
| prph2b | 0.228 | gadd45ab | -0.019 |
| tmem244 | 0.226 | mir205 | -0.019 |
| prph2a | 0.220 | pitpnaa | -0.019 |
| zgc:109965 | 0.220 | MCMDC2 | -0.019 |
| tmx3a | 0.212 | edem1 | -0.018 |
| arl3l1 | 0.211 | slc1a3a | -0.018 |
| ehd2a | 0.209 | si:ch211-76l23.4 | -0.018 |
| CU695117.1 | 0.208 | tmem39a | -0.018 |
| opn1mw1 | 0.208 | yif1a | -0.018 |
| rcvrn2 | 0.208 | wasf2 | -0.018 |
| arr3a | 0.202 | tgfb1a | -0.018 |
| arl3l2 | 0.200 | senp2 | -0.018 |
| gnat2 | 0.197 | zgc:111983 | -0.018 |
| ca7 | 0.195 | eppk1 | -0.018 |
| aipl2 | 0.194 | smco4 | -0.018 |
| slc25a3a | 0.193 | tmem167a | -0.018 |