"GLI pathogenesis-related 2, like"
ZFIN 
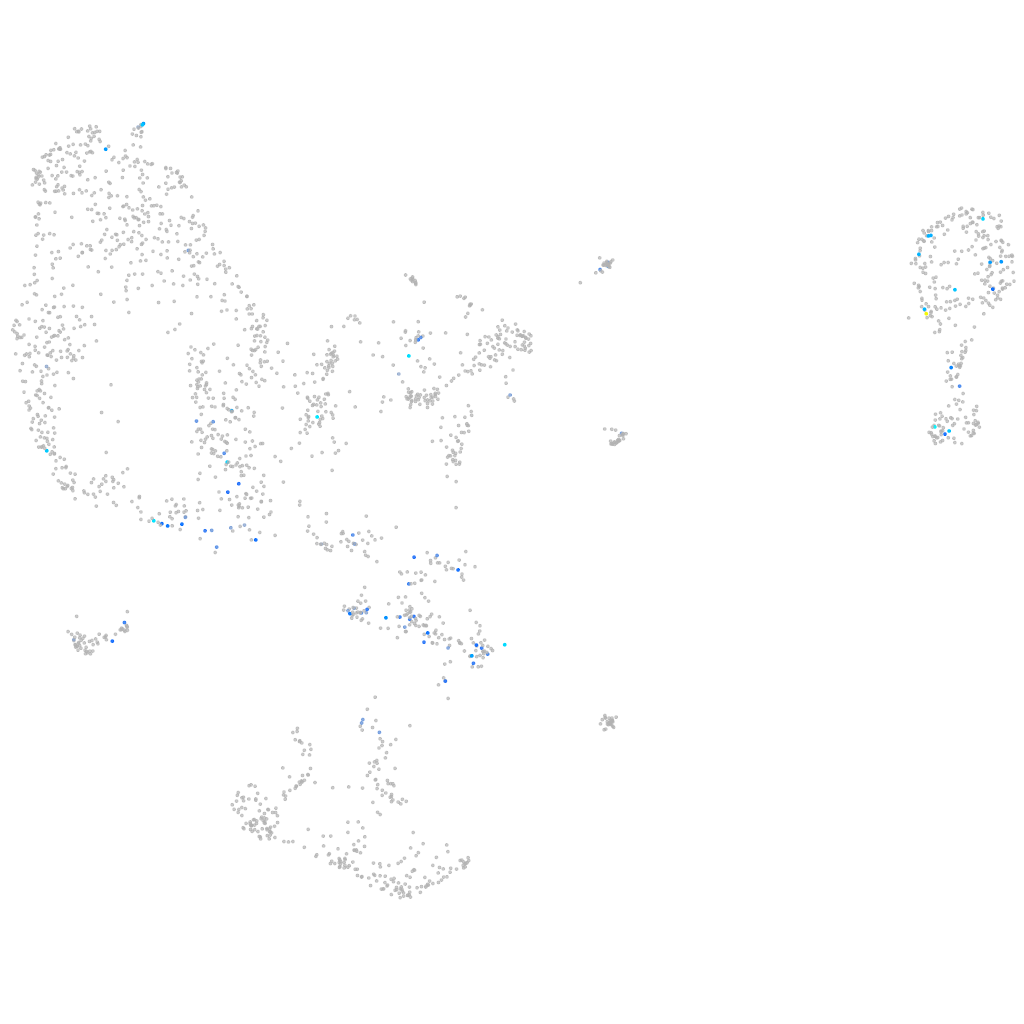
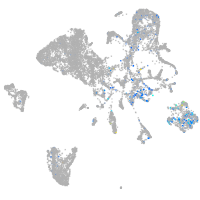
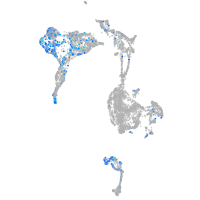
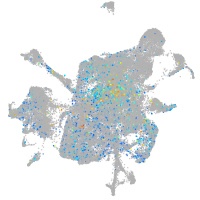
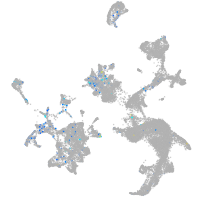
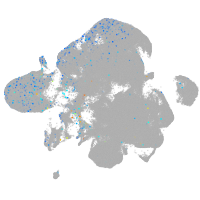
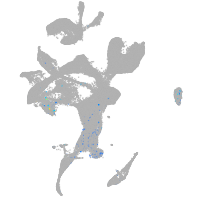
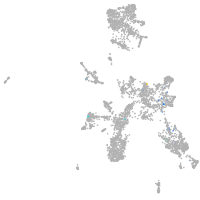
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| RHOU | 0.273 | gstp1 | -0.132 |
| zgc:77197 | 0.270 | prdx2 | -0.121 |
| nkx1.2la | 0.246 | si:ch211-139a5.9 | -0.116 |
| itln3 | 0.238 | cox7a1 | -0.111 |
| zgc:171704 | 0.233 | sod2 | -0.110 |
| tlr5a | 0.210 | cox6a2 | -0.104 |
| FO704858.1 | 0.204 | sod1 | -0.102 |
| BX545856.1 | 0.191 | gstt1a | -0.097 |
| fem1a | 0.191 | ldhba | -0.097 |
| FO393424.2 | 0.188 | glud1b | -0.096 |
| tmem144b | 0.188 | si:dkey-16p21.8 | -0.096 |
| XLOC-042955 | 0.185 | mt-atp6 | -0.095 |
| trim35-1 | 0.182 | gapdh | -0.094 |
| senp6b | 0.181 | adh5 | -0.093 |
| AL645691.1 | 0.175 | eci2 | -0.092 |
| XLOC-018699 | 0.173 | mif | -0.091 |
| LOC101885297 | 0.172 | cat | -0.091 |
| LO018166.1 | 0.172 | acadl | -0.090 |
| zbtb16a | 0.170 | aldh6a1 | -0.090 |
| anos1b | 0.168 | phyhd1 | -0.089 |
| atmin | 0.166 | atp5f1b | -0.089 |
| kcna2a | 0.166 | IYD | -0.089 |
| hif1aa | 0.166 | mt-co2 | -0.088 |
| CABZ01056843.1 | 0.163 | gcshb | -0.088 |
| LOC110437938 | 0.160 | suclg2 | -0.088 |
| LOC101886167 | 0.158 | eno3 | -0.088 |
| tespa1 | 0.157 | pvalb1 | -0.088 |
| LOC101884739 | 0.156 | eef1da | -0.088 |
| LOC108180723 | 0.156 | ndufa3 | -0.086 |
| XLOC-039646 | 0.156 | rps27l | -0.086 |
| chsy3 | 0.155 | cdaa | -0.086 |
| esyt2b | 0.154 | dpys | -0.086 |
| tcap | 0.153 | cox5b2 | -0.085 |
| LOC101882122 | 0.153 | atp5mf | -0.085 |
| si:ch211-113a14.9 | 0.152 | suclg1 | -0.085 |