chondroitin sulfate synthase 3
ZFIN 
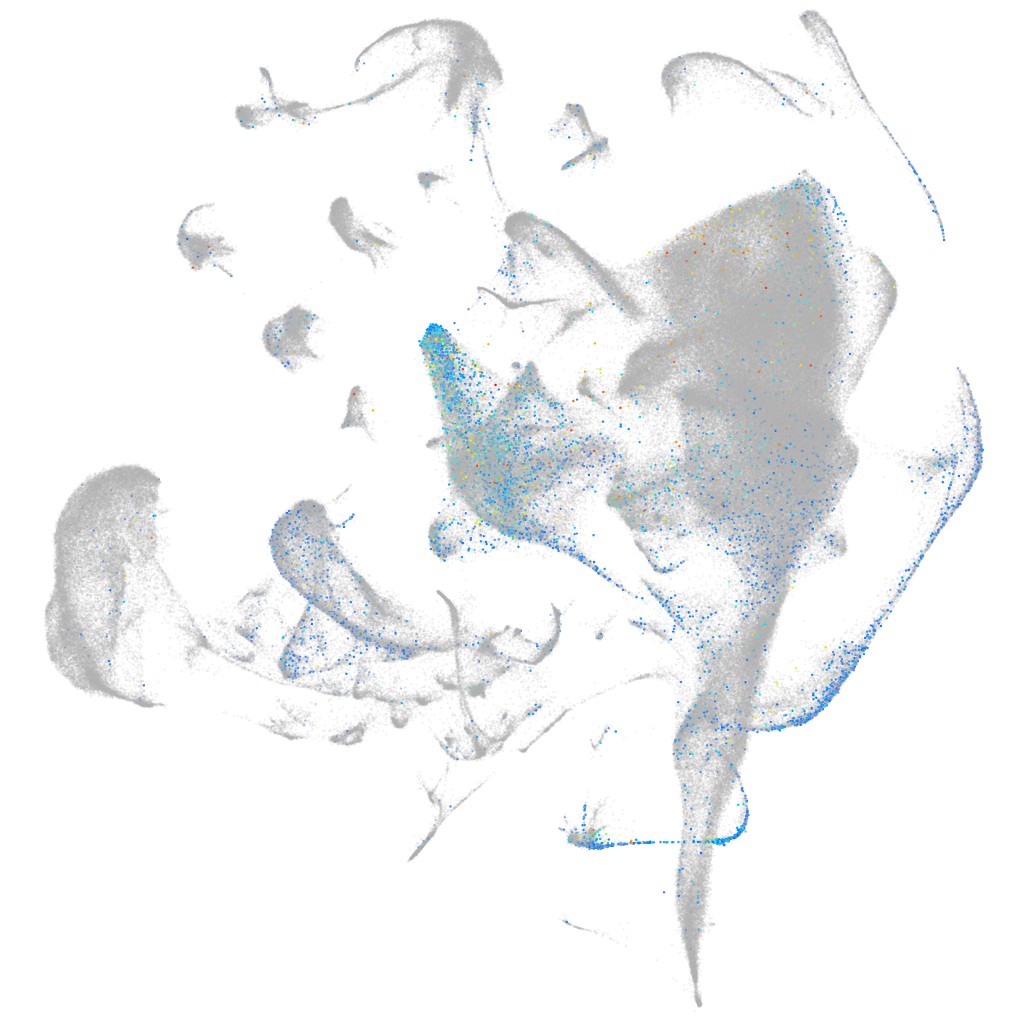
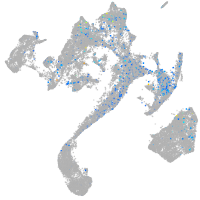
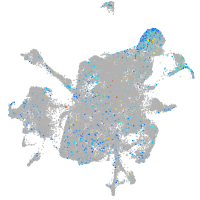
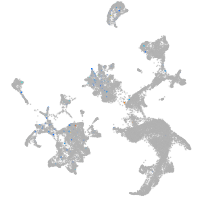
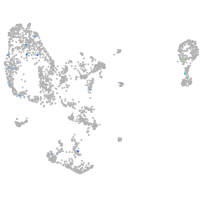
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| col9a3 | 0.157 | si:ch211-137a8.4 | -0.042 |
| cnmd | 0.155 | nova2 | -0.039 |
| col9a2 | 0.152 | fabp3 | -0.036 |
| rcn3 | 0.148 | tuba1a | -0.036 |
| col2a1a | 0.146 | tuba1c | -0.036 |
| col11a2 | 0.145 | gpm6aa | -0.034 |
| snorc | 0.145 | elavl3 | -0.033 |
| acana | 0.143 | rtn1a | -0.030 |
| emilin3a | 0.141 | sox11b | -0.030 |
| col2a1b | 0.139 | her15.1 | -0.029 |
| fgfbp2b | 0.138 | icn | -0.028 |
| fkbp14 | 0.137 | icn2 | -0.028 |
| wisp3 | 0.136 | si:ch211-195b11.3 | -0.028 |
| col11a1a | 0.134 | sox3 | -0.028 |
| kdelr3 | 0.134 | cldnb | -0.027 |
| tnxba | 0.134 | epb41a | -0.027 |
| fkbp9 | 0.133 | hmgb1b | -0.027 |
| col9a1a | 0.131 | si:dkey-276j7.1 | -0.027 |
| srpx | 0.129 | XLOC-003692 | -0.027 |
| loxl3b | 0.129 | dlb | -0.026 |
| serpinh1b | 0.128 | LOC798783 | -0.026 |
| mia | 0.127 | mgst1.2 | -0.026 |
| fkbp7 | 0.126 | myt1a | -0.026 |
| matn1 | 0.126 | rcc2 | -0.026 |
| crtap | 0.125 | stmn1b | -0.026 |
| fkbp10b | 0.122 | tmsb | -0.026 |
| csgalnact1a | 0.121 | XLOC-003689 | -0.026 |
| hapln1b | 0.120 | zgc:193505 | -0.026 |
| plod1a | 0.119 | agr1 | -0.025 |
| acanb | 0.117 | anxa1c | -0.025 |
| plod3 | 0.117 | apex1 | -0.025 |
| tmem45a | 0.117 | chd4a | -0.025 |
| zgc:153675 | 0.117 | hmgb3b | -0.025 |
| AL929222.2 | 0.116 | lye | -0.025 |
| tgfb2 | 0.116 | marcksb | -0.025 |