gdnf family receptor alpha 1a
ZFIN 
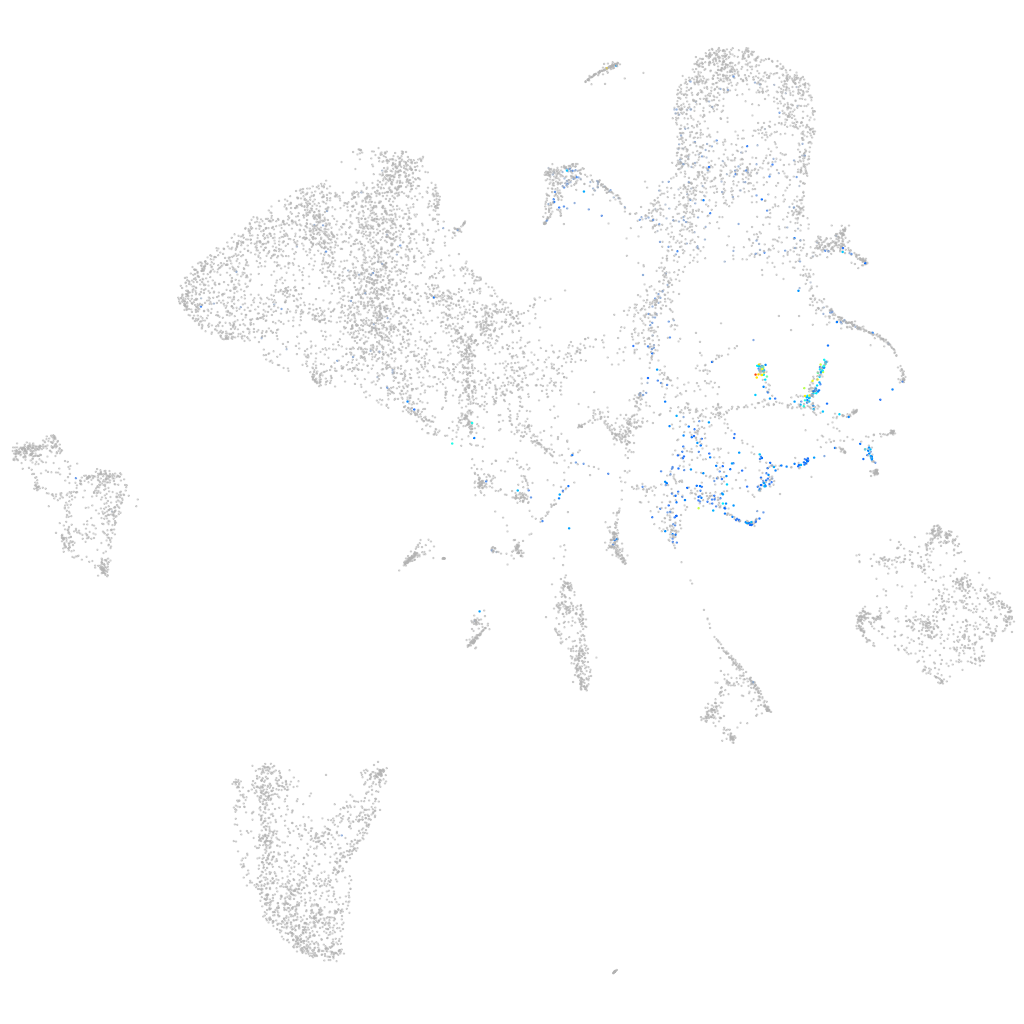
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccka | 0.433 | ahcy | -0.167 |
| si:zfos-2372e4.1 | 0.380 | gamt | -0.161 |
| scg3 | 0.339 | gapdh | -0.159 |
| rem2 | 0.316 | bhmt | -0.150 |
| rasd4 | 0.303 | gatm | -0.150 |
| pcsk1nl | 0.298 | mat1a | -0.136 |
| egr4 | 0.297 | fbp1b | -0.129 |
| insm1b | 0.289 | apoa1b | -0.125 |
| capsla | 0.286 | apoa4b.1 | -0.125 |
| pcsk2 | 0.281 | agxtb | -0.124 |
| plcxd3 | 0.280 | aldh6a1 | -0.123 |
| mir7a-1 | 0.279 | glud1b | -0.121 |
| c2cd4a | 0.277 | grhprb | -0.121 |
| scgn | 0.276 | afp4 | -0.120 |
| tspan7 | 0.275 | fabp3 | -0.120 |
| vat1 | 0.273 | tfa | -0.119 |
| fev | 0.268 | agxta | -0.117 |
| neurod1 | 0.266 | apoc2 | -0.117 |
| pax4 | 0.265 | nupr1b | -0.116 |
| mctp2b | 0.264 | cx32.3 | -0.116 |
| arxa | 0.263 | kng1 | -0.112 |
| LO018188.1 | 0.260 | apobb.1 | -0.112 |
| si:ch73-160i9.2 | 0.260 | serpina1 | -0.112 |
| trmt9b | 0.258 | serpina1l | -0.111 |
| insl5a | 0.257 | pnp4b | -0.110 |
| FADS6 | 0.254 | scp2a | -0.110 |
| s100a1 | 0.254 | apoc1 | -0.110 |
| TCIM (1 of many) | 0.251 | rbp2b | -0.110 |
| ret | 0.250 | apoa2 | -0.110 |
| onecut1 | 0.250 | ttc36 | -0.110 |
| ptger3 | 0.247 | gnmt | -0.109 |
| cckb | 0.246 | mgst1.2 | -0.109 |
| rprmb | 0.245 | fetub | -0.108 |
| pax6b | 0.243 | hao1 | -0.108 |
| calm1b | 0.242 | apom | -0.108 |