GDP dissociation inhibitor 1
ZFIN 
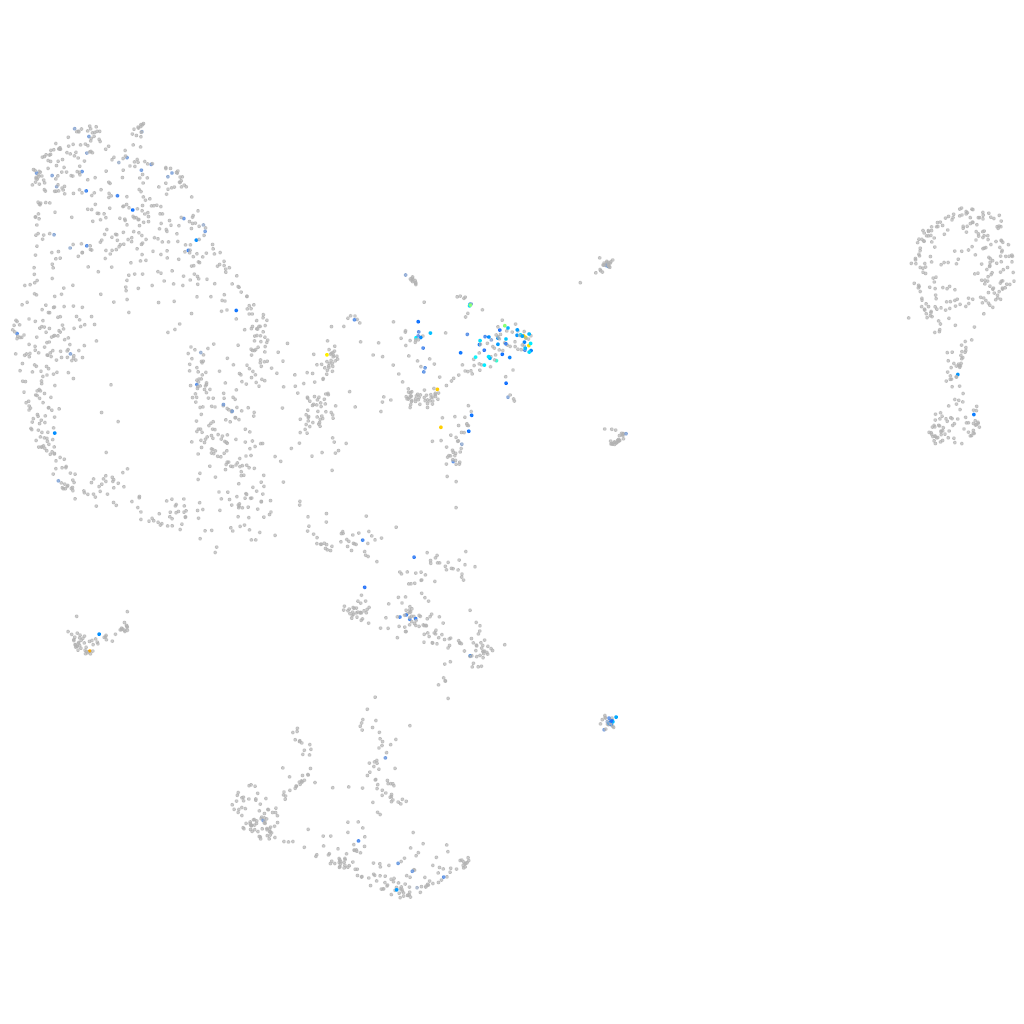
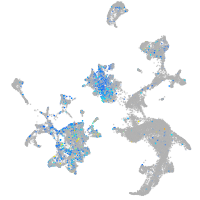
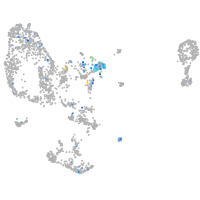
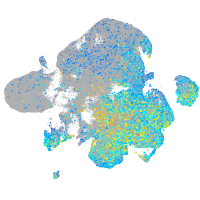
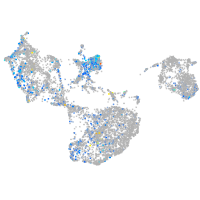
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1b | 0.425 | atp1b1a | -0.140 |
| si:dkeyp-75h12.5 | 0.424 | cdh17 | -0.120 |
| rtn1a | 0.412 | cox7b | -0.119 |
| stmn1b | 0.411 | atp1a1a.4 | -0.117 |
| mllt11 | 0.410 | sgk2a | -0.117 |
| gap43 | 0.408 | sgk1 | -0.117 |
| cdk5r1b | 0.406 | ezra | -0.115 |
| vamp2 | 0.393 | hnf1ba | -0.114 |
| gng3 | 0.389 | si:dkey-151g10.6 | -0.112 |
| sncb | 0.388 | rpl17 | -0.108 |
| nova2 | 0.386 | atp5if1a | -0.108 |
| palm1b | 0.385 | rpl4 | -0.107 |
| maptb | 0.383 | etnk1 | -0.105 |
| myt1la | 0.383 | sypl2a | -0.103 |
| gpm6aa | 0.383 | NC-002333.4 | -0.102 |
| tuba1c | 0.382 | uqcrc2b | -0.102 |
| stxbp1a | 0.382 | c1qbp | -0.101 |
| tubb5 | 0.372 | spint2 | -0.101 |
| nptnb | 0.370 | clic5b | -0.100 |
| ckbb | 0.368 | tmprss2 | -0.099 |
| ywhag2 | 0.367 | ca2 | -0.098 |
| eno2 | 0.367 | ccng1 | -0.098 |
| stmn2a | 0.364 | cd9b | -0.097 |
| stx1b | 0.361 | si:ch211-195e19.1 | -0.097 |
| dnajc5aa | 0.361 | f11r.1 | -0.096 |
| celf5a | 0.359 | slc26a6 | -0.096 |
| gabrb4 | 0.355 | rbm47 | -0.096 |
| csdc2a | 0.355 | ndrg1a | -0.096 |
| si:ch211-214j24.9 | 0.354 | pdzk1 | -0.096 |
| napgb | 0.352 | rnaseka | -0.095 |
| gpm6ab | 0.351 | exoc3l2b | -0.095 |
| rac3a | 0.351 | aldh6a1 | -0.094 |
| mogat2 | 0.350 | pttg1ipb | -0.094 |
| atp6v0cb | 0.347 | cox7c | -0.094 |
| sprn | 0.345 | rpl32 | -0.093 |