GDP dissociation inhibitor 1
ZFIN 
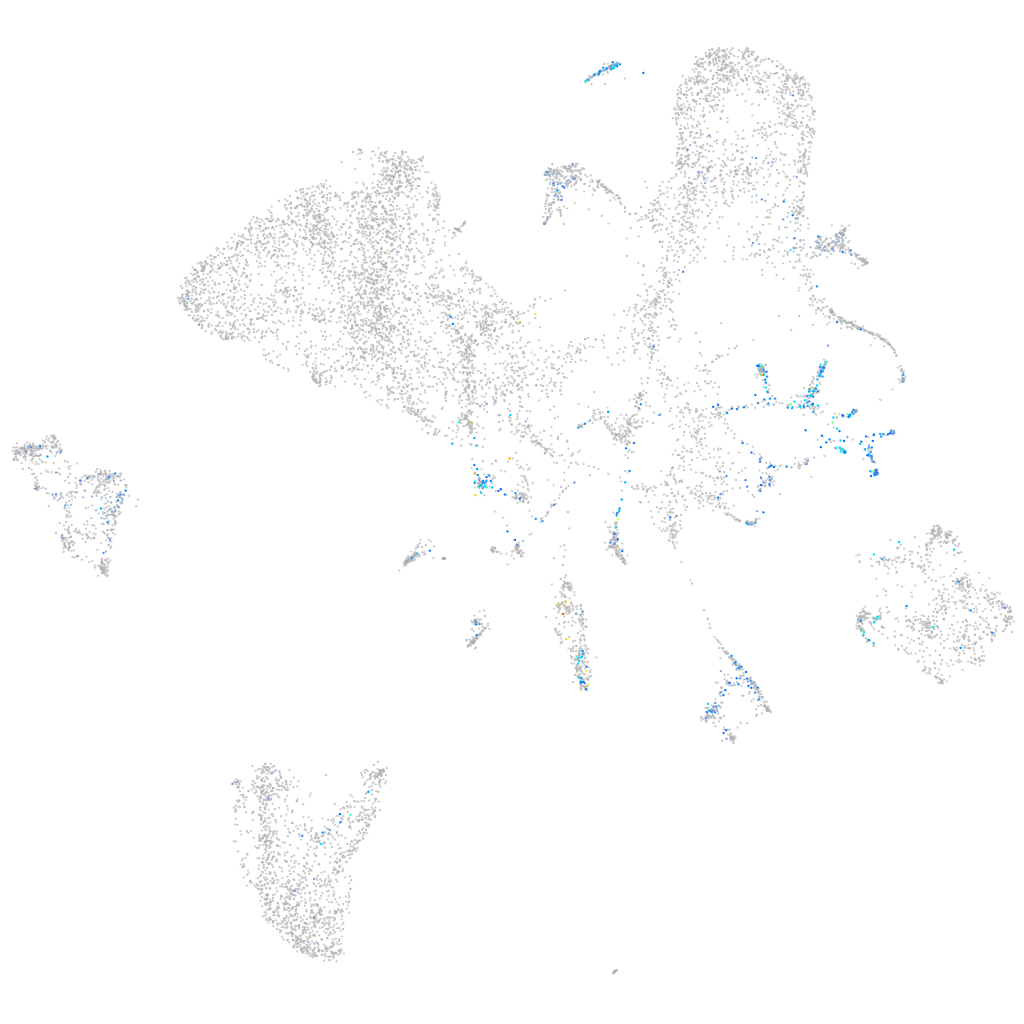
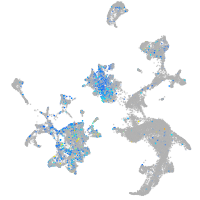
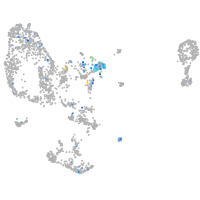
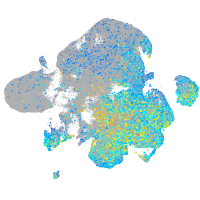
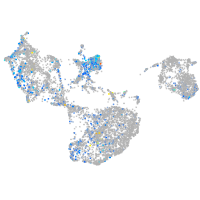
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.436 | gapdh | -0.179 |
| neurod1 | 0.386 | ahcy | -0.177 |
| pcsk1nl | 0.382 | aldh6a1 | -0.167 |
| mir7a-1 | 0.361 | gamt | -0.163 |
| dpysl2b | 0.338 | mat1a | -0.157 |
| vat1 | 0.334 | gstt1a | -0.157 |
| syt1a | 0.333 | gstr | -0.156 |
| vamp2 | 0.333 | aldh7a1 | -0.155 |
| egr4 | 0.330 | aldob | -0.152 |
| pax6b | 0.327 | scp2a | -0.150 |
| atp6v0cb | 0.324 | agxta | -0.148 |
| insm1b | 0.322 | cx32.3 | -0.148 |
| insm1a | 0.312 | agxtb | -0.145 |
| scgn | 0.312 | fbp1b | -0.142 |
| gapdhs | 0.311 | fabp3 | -0.142 |
| zgc:101731 | 0.309 | gatm | -0.141 |
| fev | 0.307 | glud1b | -0.140 |
| rasd4 | 0.307 | gcshb | -0.140 |
| rnasekb | 0.305 | nipsnap3a | -0.139 |
| lysmd2 | 0.303 | apoa4b.1 | -0.139 |
| gpc1a | 0.302 | hspe1 | -0.138 |
| gnao1a | 0.300 | mgst1.2 | -0.138 |
| id4 | 0.300 | ftcd | -0.138 |
| atpv0e2 | 0.298 | pnp4b | -0.137 |
| tspan7b | 0.295 | aqp12 | -0.137 |
| pcsk1 | 0.294 | apoa1b | -0.136 |
| si:dkey-153k10.9 | 0.292 | eno3 | -0.135 |
| kcnj11 | 0.290 | nupr1b | -0.133 |
| calm1b | 0.288 | fabp10a | -0.132 |
| nkx2.2a | 0.286 | hspd1 | -0.131 |
| hsbp1a | 0.280 | tdo2a | -0.131 |
| tmsb | 0.279 | ttc36 | -0.131 |
| hepacam2 | 0.276 | sult2st2 | -0.131 |
| myt1b | 0.276 | aldh1l1 | -0.131 |
| mllt11 | 0.275 | bhmt | -0.131 |