growth differentiation factor 10b
ZFIN 
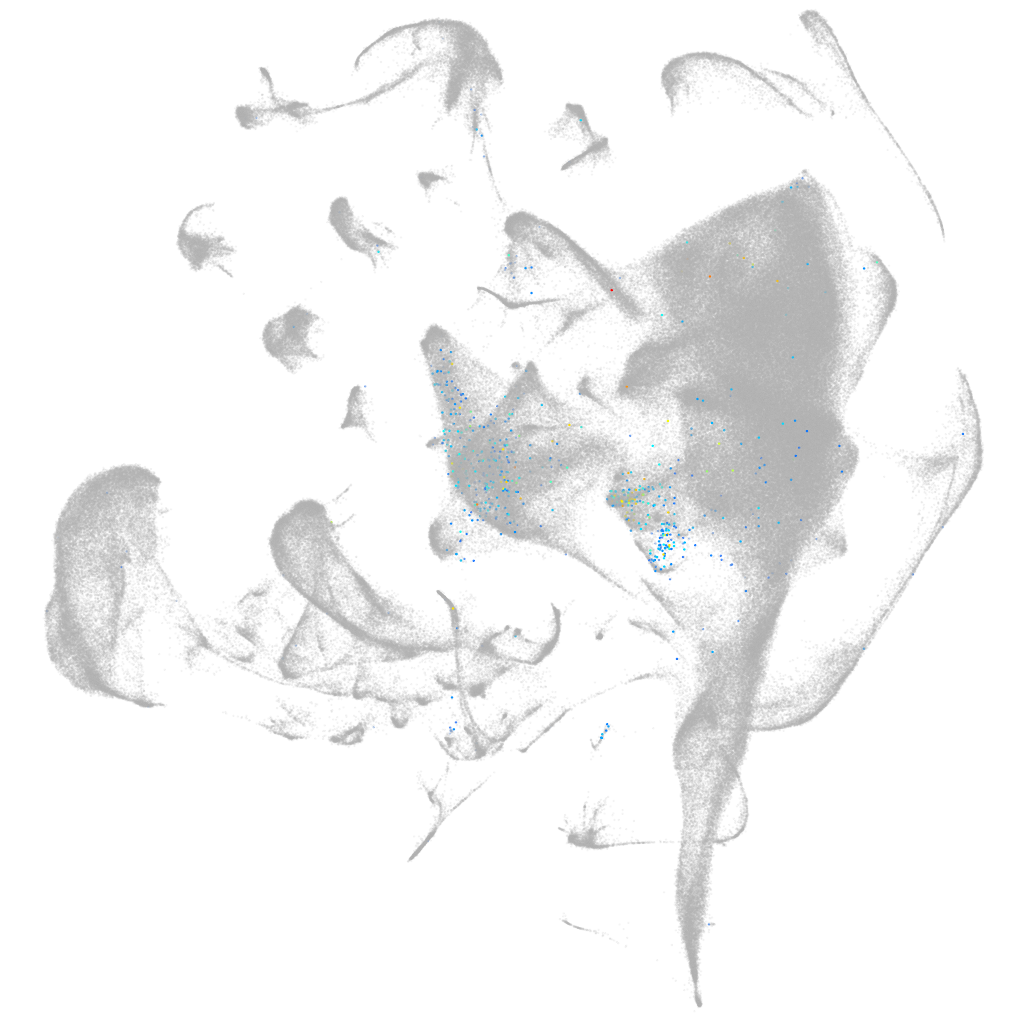
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPATA1 | 0.104 | apex1 | -0.013 |
| hoxb13a | 0.086 | cfl1l | -0.013 |
| si:dkey-183j2.10 | 0.086 | epcam | -0.013 |
| c8h22orf15 | 0.081 | hmgb3b | -0.013 |
| si:ch211-250c4.5 | 0.073 | ilf2 | -0.013 |
| hoxc13b | 0.071 | marcksb | -0.013 |
| pald1b | 0.071 | cyt1 | -0.012 |
| hoxc13a | 0.065 | hnrnpa1a | -0.012 |
| si:dkeyp-69c1.9 | 0.064 | hnrnpub | -0.012 |
| dthd1 | 0.063 | krt4 | -0.012 |
| si:ch211-66e2.5 | 0.063 | sox11b | -0.012 |
| hoxa13a | 0.061 | cd9b | -0.011 |
| si:dkeyp-110a12.4 | 0.061 | cycsb | -0.011 |
| LOC100329490 | 0.059 | cyt1l | -0.011 |
| BX469902.1 | 0.058 | smarca4a | -0.011 |
| gdf10a | 0.057 | tmsb1 | -0.011 |
| si:ch211-155m12.5 | 0.057 | wu:fb18f06 | -0.011 |
| cfap157 | 0.056 | abracl | -0.010 |
| prss35 | 0.056 | cdh1 | -0.010 |
| spata18 | 0.055 | ddx39ab | -0.010 |
| s100b | 0.054 | gsnb | -0.010 |
| sept8b | 0.053 | icn | -0.010 |
| tbata | 0.053 | lyar | -0.010 |
| ccdc169 | 0.051 | lye | -0.010 |
| mxra8b | 0.050 | mphosph10 | -0.010 |
| mfap2 | 0.050 | myt1b | -0.010 |
| nck1a | 0.049 | nop58 | -0.010 |
| si:ch211-163l21.7 | 0.049 | npm1a | -0.010 |
| smkr1 | 0.049 | oclna | -0.010 |
| hoxa13b | 0.048 | prr13 | -0.010 |
| tppp3 | 0.048 | pycard | -0.010 |
| cdkl1 | 0.047 | si:ch73-21g5.7 | -0.010 |
| ccdc175 | 0.046 | snrpd1 | -0.010 |
| si:dkey-7j14.6 | 0.046 | top1l | -0.010 |
| slc1a3b | 0.046 | zgc:153284 | -0.010 |