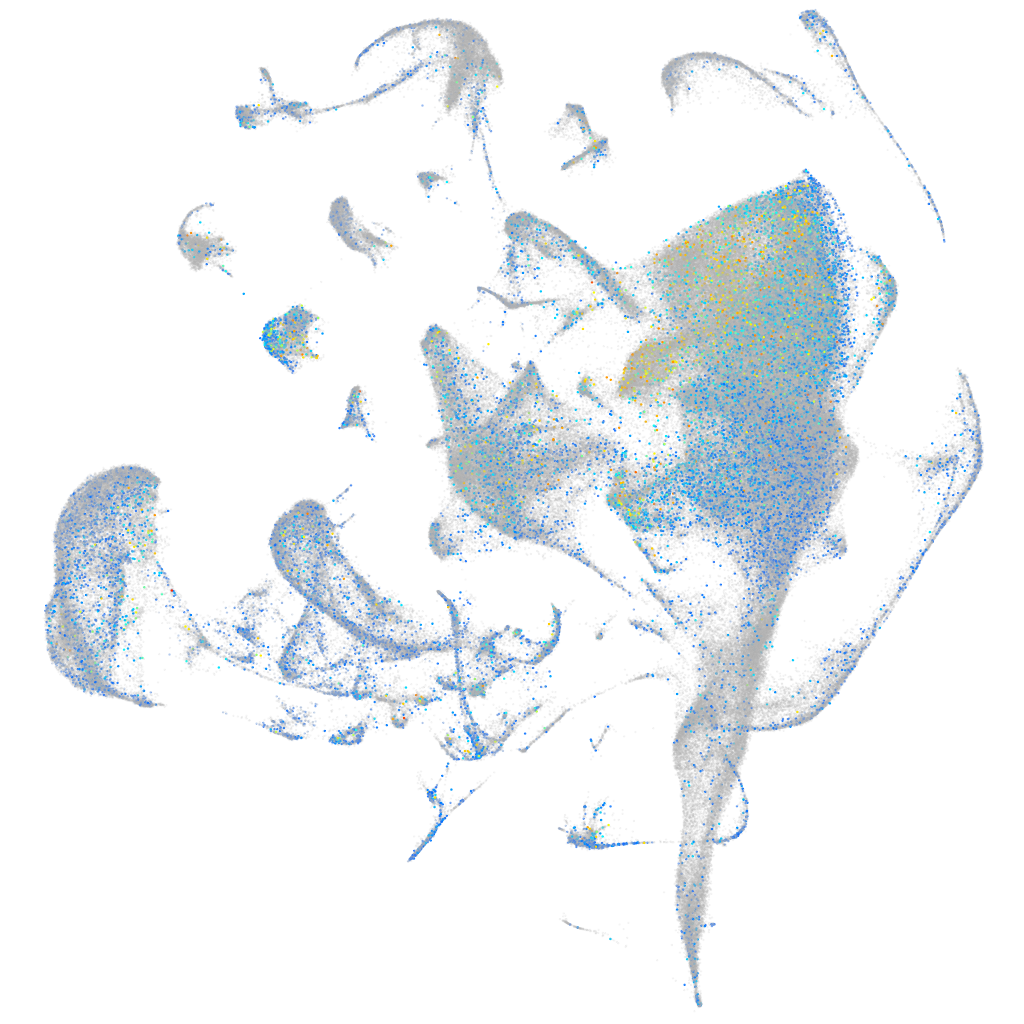GTPase activating protein and VPS9 domains 1
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nova2 | 0.050 | hspb1 | -0.042 |
| elavl3 | 0.049 | npm1a | -0.038 |
| tuba1c | 0.049 | dkc1 | -0.037 |
| gnb1a | 0.047 | nop58 | -0.036 |
| ywhah | 0.047 | apoc1 | -0.035 |
| fam168a | 0.045 | polr3gla | -0.035 |
| dpysl2b | 0.045 | pou5f3 | -0.035 |
| marcksl1a | 0.045 | si:ch211-152c2.3 | -0.035 |
| tuba1a | 0.044 | fbl | -0.034 |
| calm1a | 0.044 | rsl1d1 | -0.034 |
| celf2 | 0.043 | zmp:0000000624 | -0.034 |
| dpysl3 | 0.043 | si:dkey-66i24.9 | -0.032 |
| si:ch1073-429i10.3.1 | 0.043 | bms1 | -0.031 |
| gnao1a | 0.042 | cdca7a | -0.031 |
| gpm6aa | 0.042 | gar1 | -0.031 |
| myt1b | 0.042 | nop56 | -0.031 |
| stmn1b | 0.042 | s100a1 | -0.031 |
| myt1a | 0.041 | stm | -0.031 |
| tubb5 | 0.041 | ved | -0.031 |
| mbd3b | 0.041 | vox | -0.031 |
| gabarapb | 0.040 | zgc:56699 | -0.031 |
| stx1b | 0.040 | COX7A2 | -0.030 |
| mrc1a | 0.040 | ddx18 | -0.030 |
| ccni | 0.039 | gnl3 | -0.030 |
| gng2 | 0.039 | lig1 | -0.030 |
| hmgb3a | 0.039 | nop2 | -0.030 |
| jpt1b | 0.039 | asb11 | -0.029 |
| rab2a | 0.039 | kri1 | -0.029 |
| rtn1a | 0.039 | lrwd1 | -0.029 |
| vat1 | 0.039 | si:ch1073-80i24.3 | -0.029 |
| kif5bb | 0.039 | tbx16 | -0.029 |
| appa | 0.038 | ebna1bp2 | -0.028 |
| dbn1 | 0.038 | ing5b | -0.028 |
| elavl4 | 0.038 | ISCU | -0.028 |
| gatad2b | 0.038 | LOC108190024 | -0.028 |