GRB2-associated binding protein 1
ZFIN 
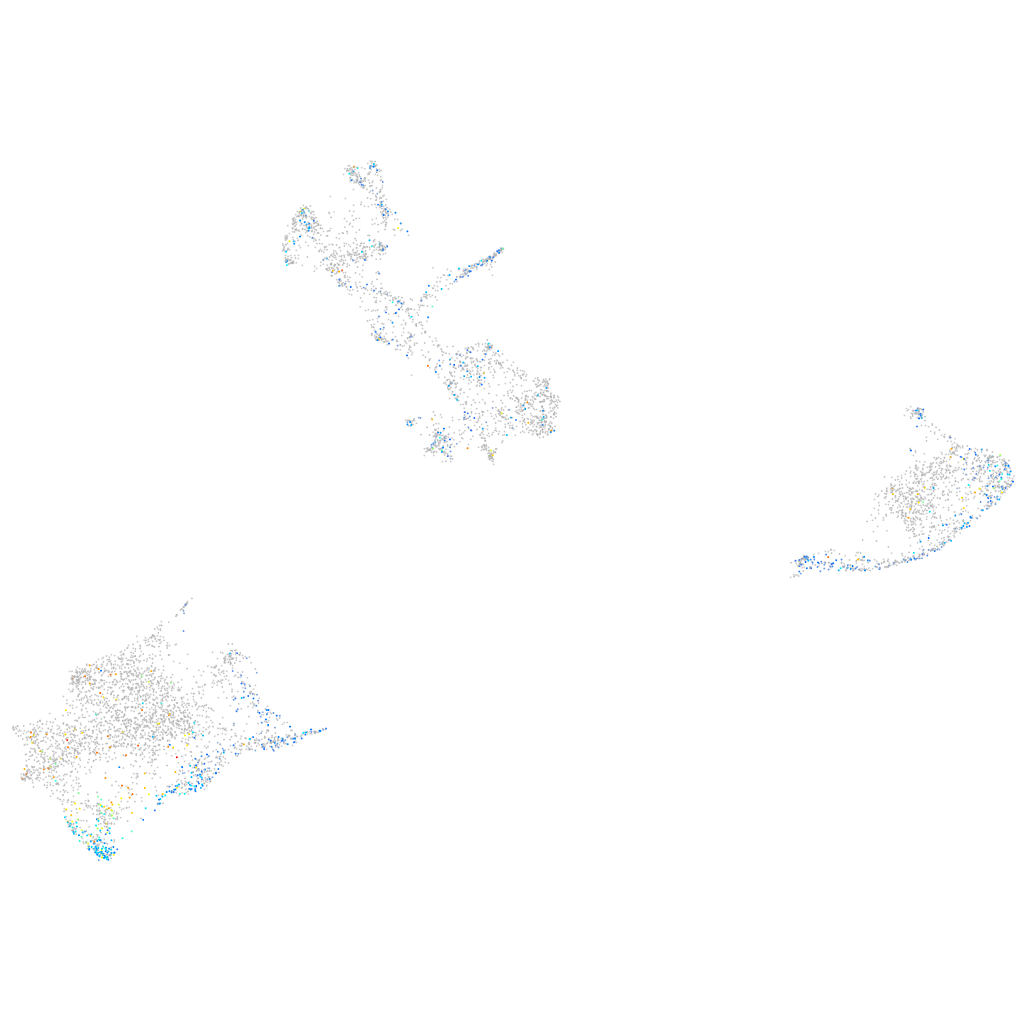
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sik1 | 0.185 | tubb2b | -0.085 |
| inka1b | 0.183 | rps12 | -0.075 |
| rasd1 | 0.174 | ptmab | -0.069 |
| trpm1b | 0.170 | si:ch211-222l21.1 | -0.063 |
| dio3a | 0.164 | ptmaa | -0.062 |
| noctb | 0.160 | hnrnpaba | -0.062 |
| wsb1 | 0.159 | h3f3a | -0.061 |
| socs3a | 0.157 | tuba1c | -0.058 |
| gadd45bb | 0.157 | rpl19 | -0.057 |
| pax7b | 0.157 | elavl3 | -0.056 |
| sdc4 | 0.149 | tubb5 | -0.053 |
| CR383676.1 | 0.148 | hmgn2 | -0.053 |
| si:zfos-1069f5.1 | 0.143 | atp5mc1 | -0.053 |
| pim1 | 0.143 | mdkb | -0.051 |
| crema | 0.140 | rps11 | -0.051 |
| f3b | 0.138 | rps15 | -0.051 |
| runx3 | 0.137 | nova2 | -0.051 |
| pnp5a | 0.136 | hbae1.1 | -0.051 |
| itpkb | 0.135 | slc25a5 | -0.050 |
| pxk | 0.135 | gng3 | -0.050 |
| XLOC-004408 | 0.134 | rplp2l | -0.050 |
| vps9d1 | 0.134 | prdx1 | -0.050 |
| zgc:92161 | 0.133 | gapdhs | -0.048 |
| plin6 | 0.131 | CU634008.1 | -0.047 |
| CR847566.1 | 0.130 | tuba1a | -0.046 |
| socs1a | 0.126 | tmsb | -0.046 |
| CABZ01072614.1 | 0.126 | rnasekb | -0.045 |
| cebpd | 0.125 | rpl39 | -0.045 |
| sesn1 | 0.125 | hmgb2b | -0.045 |
| rgrb | 0.125 | gpm6aa | -0.045 |
| kank2 | 0.125 | ywhag2 | -0.045 |
| lipeb | 0.125 | rpl21 | -0.045 |
| apol1 | 0.124 | meis2a | -0.044 |
| XLOC-002533 | 0.123 | phgdh | -0.044 |
| opn5 | 0.123 | hmgn3 | -0.044 |