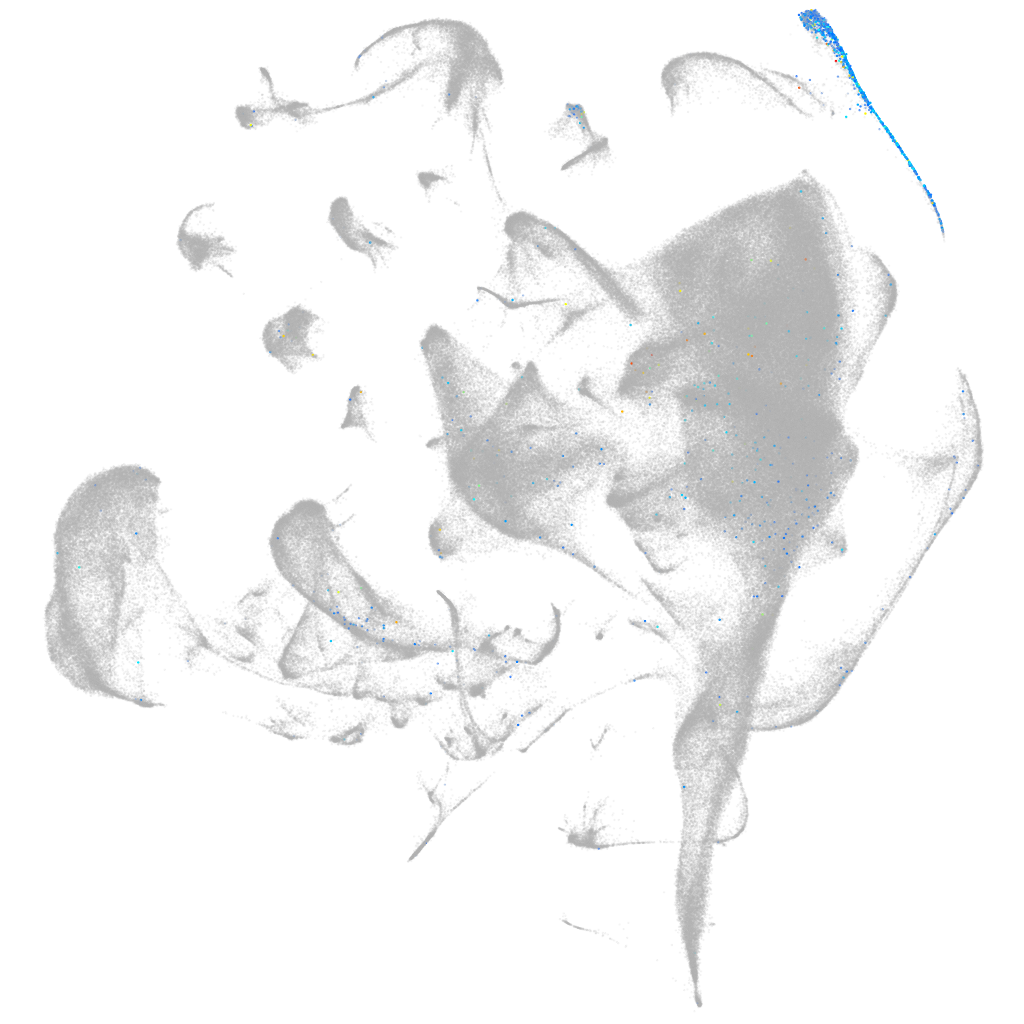
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm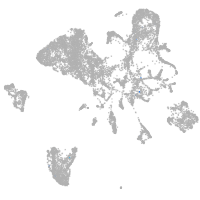 |
axial mesoderm |
muscle 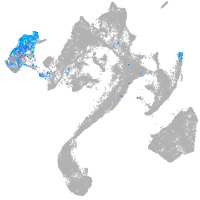 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 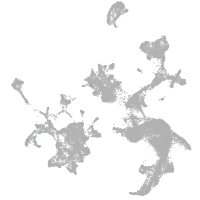 |
pronephros 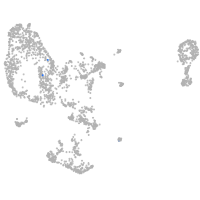 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis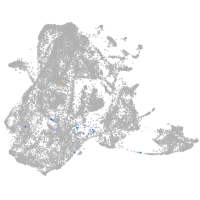 |
periderm |
fin |
ionocytes / mucous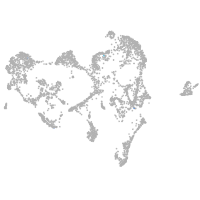 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU633479.2 | 0.511 | gpm6aa | -0.047 |
| myoz2b | 0.467 | ckbb | -0.046 |
| ryr1a | 0.463 | gpm6ab | -0.046 |
| tnni1c | 0.461 | atp6v0cb | -0.045 |
| LO018250.1 | 0.456 | gapdhs | -0.045 |
| smyhc2 | 0.455 | rnasekb | -0.045 |
| myh7bb | 0.443 | tuba1c | -0.044 |
| smyhc3 | 0.440 | ywhaqb | -0.039 |
| si:ch211-28p3.4 | 0.434 | atpv0e2 | -0.038 |
| klhl41a | 0.421 | mdkb | -0.038 |
| smyhc1 | 0.421 | nova2 | -0.038 |
| zgc:86709 | 0.421 | marcksl1a | -0.038 |
| mustn1b | 0.406 | hmgb1b | -0.037 |
| hhatlb | 0.401 | sncb | -0.037 |
| tnnc1b | 0.394 | stmn1b | -0.036 |
| myl13 | 0.392 | vamp2 | -0.036 |
| LOC103910269 | 0.391 | calm1a | -0.036 |
| myl10 | 0.391 | cspg5a | -0.035 |
| kcnmb2 | 0.381 | sypb | -0.035 |
| tnnt2c | 0.374 | atp1a3a | -0.034 |
| mylk4a | 0.361 | chd4a | -0.034 |
| myom1b | 0.358 | rtn1b | -0.034 |
| tnni4b.2 | 0.351 | tubb5 | -0.034 |
| hspb11 | 0.350 | zc4h2 | -0.034 |
| rnf207b | 0.349 | cadm3 | -0.033 |
| smpx | 0.349 | calm2a | -0.033 |
| srl | 0.344 | gng3 | -0.033 |
| chrnb1l | 0.340 | pvalb2 | -0.033 |
| flnca | 0.339 | stx1b | -0.033 |
| alpk3b | 0.338 | atp6ap2 | -0.032 |
| tnni1d | 0.337 | cpe | -0.032 |
| zgc:158296 | 0.337 | elavl3 | -0.032 |
| apobec2a | 0.334 | elavl4 | -0.032 |
| myf6 | 0.326 | si:dkeyp-75h12.5 | -0.032 |
| casq2 | 0.324 | tuba1a | -0.032 |