frizzled class receptor 8b
ZFIN 
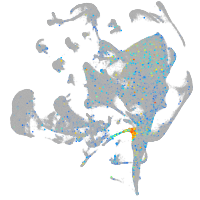
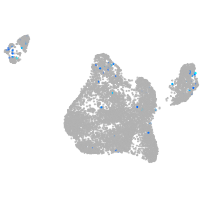
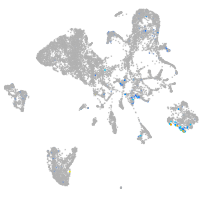
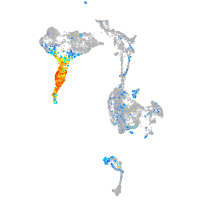
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nkx2.9 | 0.206 | syt5b | -0.078 |
| lft1 | 0.196 | crx | -0.072 |
| dact2 | 0.159 | otx5 | -0.070 |
| shisa2a | 0.158 | ckbb | -0.064 |
| fgf3 | 0.155 | sypb | -0.064 |
| hspb1 | 0.154 | rs1a | -0.063 |
| tdgf1 | 0.143 | fabp7a | -0.057 |
| sox19a | 0.140 | atp2b1b | -0.056 |
| lft2 | 0.135 | atp5mc1 | -0.056 |
| chrd | 0.133 | hsp90aa1.2 | -0.055 |
| alcamb | 0.127 | cplx4a | -0.054 |
| stm | 0.125 | neurod1 | -0.054 |
| gsc | 0.123 | tulp1a | -0.054 |
| nr6a1a | 0.122 | ppdpfa | -0.053 |
| apoeb | 0.119 | gngt2a | -0.052 |
| si:ch211-152c2.3 | 0.116 | rbp4l | -0.052 |
| fzd8a | 0.115 | ckmt2a | -0.052 |
| dnmt3bb.2 | 0.110 | elovl4b | -0.052 |
| nkx2.4b | 0.109 | LOC103909376 | -0.052 |
| ndr2 | 0.108 | neurod4 | -0.052 |
| hesx1 | 0.106 | pvalb1 | -0.051 |
| COX7A2 | 0.105 | si:ch73-28h20.1 | -0.051 |
| NC-002333.4 | 0.105 | gapdhs | -0.051 |
| aldob | 0.104 | ndrg1a | -0.050 |
| nkx2.2b | 0.103 | tmx3a | -0.049 |
| fthl27 | 0.100 | rcvrn2 | -0.049 |
| cyp26a1 | 0.100 | ddah2 | -0.049 |
| marcksb | 0.098 | nexn | -0.049 |
| ncl | 0.097 | stxbp1b | -0.049 |
| nop58 | 0.097 | pvalb2 | -0.048 |
| npm1a | 0.096 | gngt2b | -0.048 |
| si:ch211-152c8.5 | 0.093 | unc119b | -0.048 |
| bms1 | 0.093 | zgc:109965 | -0.047 |
| dkc1 | 0.092 | arl3l1 | -0.047 |
| ebna1bp2 | 0.092 | ndrg1b | -0.046 |