"fucosyltransferase 8b (alpha (1,6) fucosyltransferase)"
ZFIN 
Other cell groups


all cells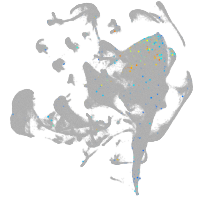 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 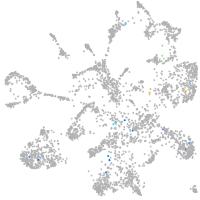 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia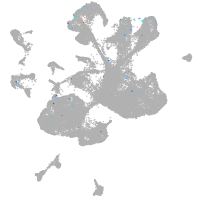 |
eye |
taste / olfactory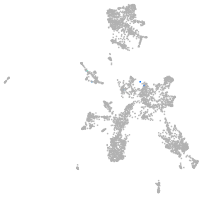 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01063034.1 | 0.115 | hmgb2a | -0.054 |
| CU914760.1 | 0.106 | tpt1 | -0.036 |
| atp2b3b | 0.105 | rps20 | -0.033 |
| gpha2 | 0.103 | rps8a | -0.032 |
| eno2 | 0.103 | hmga1a | -0.032 |
| LOC110439758 | 0.103 | rplp2l | -0.032 |
| FAM163B (1 of many) | 0.099 | rpl8 | -0.031 |
| syt2a | 0.099 | rps19 | -0.031 |
| sv2a | 0.096 | rpl11 | -0.031 |
| rasal1a | 0.094 | rpl18 | -0.031 |
| si:dkeyp-72g9.4 | 0.093 | rpl5b | -0.031 |
| gabrb4 | 0.092 | eef1a1l1 | -0.031 |
| ywhag1 | 0.090 | rack1 | -0.030 |
| BX276128.1 | 0.088 | rps6 | -0.030 |
| stmn4 | 0.088 | si:dkey-151g10.6 | -0.030 |
| grin1a | 0.088 | rps12 | -0.029 |
| npas4a | 0.086 | rps17 | -0.029 |
| snap25a | 0.084 | ahcy | -0.029 |
| oaz2b | 0.084 | rps15a | -0.029 |
| scn8aa | 0.083 | rps21 | -0.029 |
| carmil2 | 0.081 | rpl12 | -0.029 |
| vsnl1b | 0.081 | rpl22l1 | -0.028 |
| cplx2l | 0.080 | rpl39 | -0.028 |
| rbpms2a | 0.080 | rpl30 | -0.028 |
| clstn2 | 0.080 | rpl3 | -0.028 |
| elmod1 | 0.079 | rps24 | -0.028 |
| vsnl1a | 0.079 | rpl31 | -0.028 |
| CU207343.2 | 0.079 | fabp7a | -0.028 |
| chrnb3b | 0.079 | rpl15 | -0.028 |
| slc12a5b | 0.078 | rps27.1 | -0.028 |
| BX248418.1 | 0.078 | rpl10 | -0.028 |
| kcnc3b | 0.078 | rps13 | -0.028 |
| LOC101883723 | 0.077 | rps4x | -0.027 |
| LOC110438239 | 0.077 | rplp1 | -0.027 |
| stmn2a | 0.077 | rpl7a | -0.027 |




