"fascin actin-bundling protein 2b, retinal"
ZFIN 
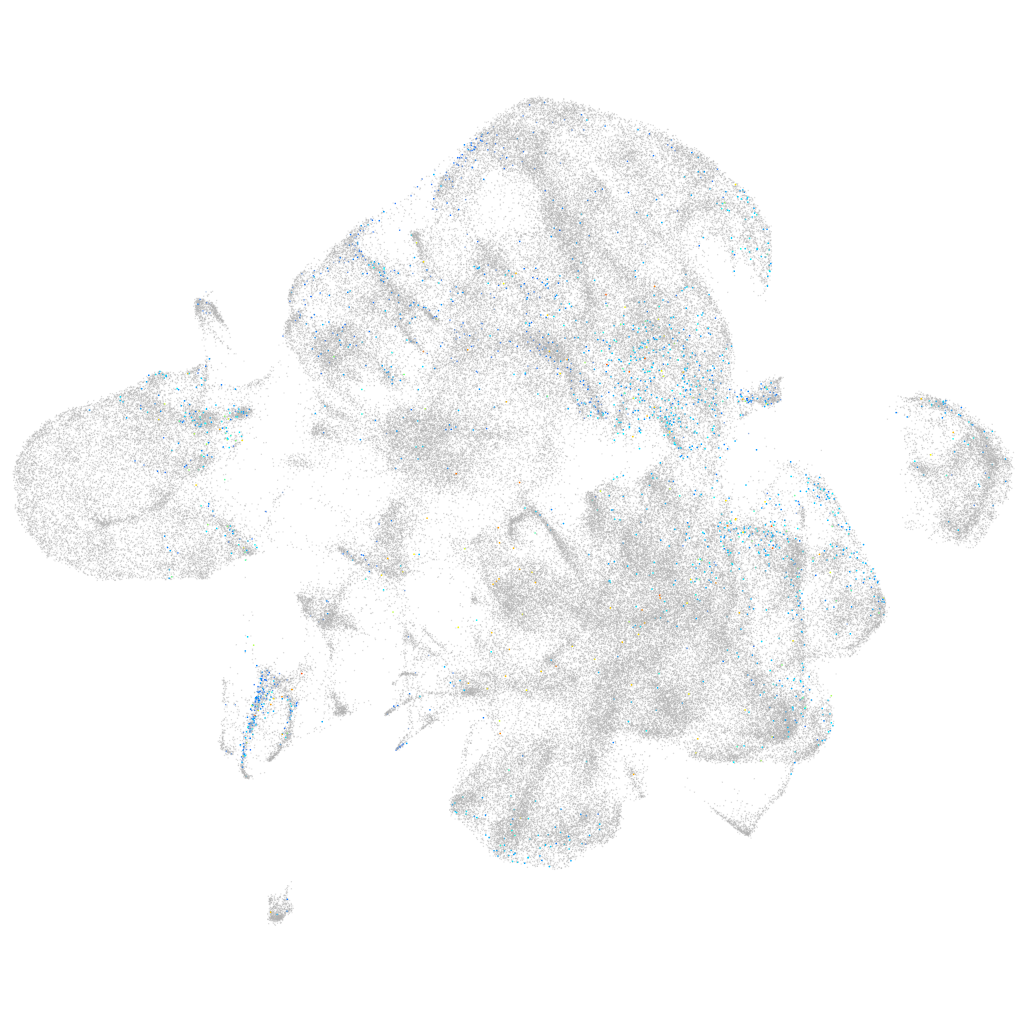
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dlb | 0.093 | cspg5a | -0.032 |
| ebf2 | 0.081 | snap25a | -0.030 |
| neurod1 | 0.071 | sv2a | -0.030 |
| im:7152348 | 0.070 | sypb | -0.028 |
| sox11b | 0.068 | syt1a | -0.028 |
| insm1a | 0.068 | ywhag2 | -0.027 |
| myt1a | 0.062 | stmn2a | -0.027 |
| tp53inp2 | 0.058 | sncb | -0.026 |
| insm1b | 0.057 | slc6a1a | -0.026 |
| neurod4 | 0.057 | spock3 | -0.025 |
| rtca | 0.057 | sncgb | -0.025 |
| scrt2 | 0.056 | vsnl1b | -0.025 |
| hes6 | 0.056 | atp2b2 | -0.025 |
| cdkn1ca | 0.055 | tmsb2 | -0.025 |
| nhlh2 | 0.055 | eno2 | -0.025 |
| elavl3 | 0.053 | stx1b | -0.025 |
| LOC798783 | 0.053 | pcna | -0.025 |
| si:dkey-56m19.5 | 0.052 | gabrb4 | -0.025 |
| dla | 0.052 | calm2b | -0.025 |
| lima1a | 0.051 | rab11bb | -0.025 |
| notch1a | 0.051 | cdk5r2a | -0.025 |
| eya4 | 0.050 | scg2b | -0.025 |
| CU634008.1 | 0.050 | ptprga | -0.024 |
| myt1b | 0.049 | tpi1b | -0.024 |
| six2b | 0.048 | syngr3a | -0.023 |
| si:ch211-222l21.1 | 0.048 | snap25b | -0.023 |
| hmgn6 | 0.047 | syn2a | -0.023 |
| peli1b | 0.047 | rtn1b | -0.023 |
| bcl2l10 | 0.046 | camk2n1a | -0.023 |
| neurod6b | 0.046 | eno1a | -0.023 |
| kdm2bb | 0.045 | calm1b | -0.023 |
| six1b | 0.044 | atp6v0cb | -0.023 |
| BX530077.1 | 0.044 | si:ch73-127m5.1 | -0.023 |
| smarcb1b | 0.044 | chd3 | -0.023 |
| si:ch211-193l2.6 | 0.043 | ywhag1 | -0.022 |