furry homolog b (Drosophila)
ZFIN 
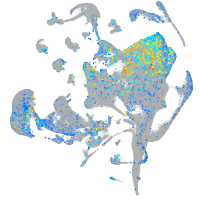
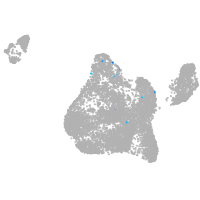
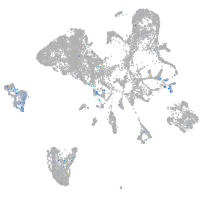
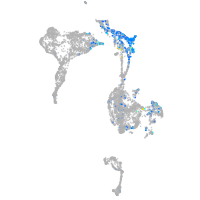
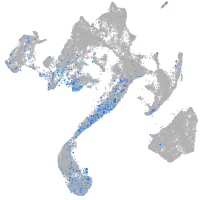
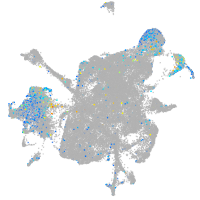
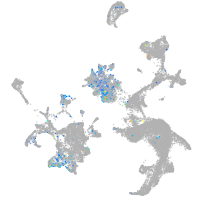
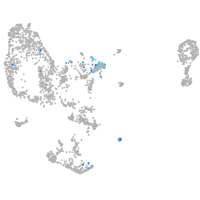
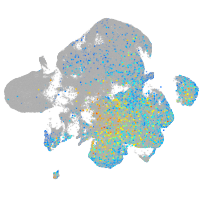
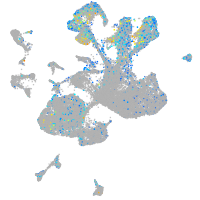
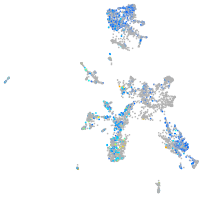
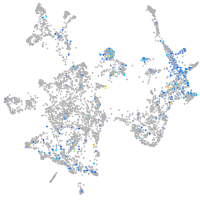
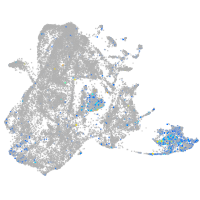
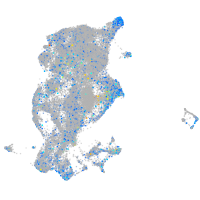
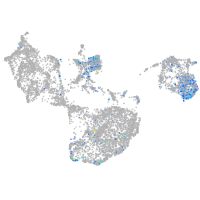
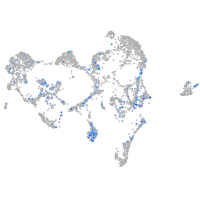
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl4 | 0.242 | XLOC-003690 | -0.174 |
| stx1b | 0.232 | id1 | -0.167 |
| snap25a | 0.227 | mdka | -0.166 |
| gng3 | 0.226 | hmgb2a | -0.163 |
| stxbp1a | 0.215 | her15.1 | -0.161 |
| syt2a | 0.210 | cldn5a | -0.156 |
| sncb | 0.210 | si:dkey-151g10.6 | -0.147 |
| tmem59l | 0.208 | sdc4 | -0.144 |
| stmn1b | 0.206 | sox3 | -0.143 |
| atp6v0cb | 0.205 | rplp1 | -0.139 |
| rtn1b | 0.204 | XLOC-003689 | -0.139 |
| cplx2 | 0.204 | msna | -0.135 |
| ywhah | 0.202 | sparc | -0.134 |
| rbfox1 | 0.201 | rpl26 | -0.133 |
| elavl3 | 0.199 | rpl12 | -0.133 |
| stmn2a | 0.198 | rps2 | -0.132 |
| ywhag2 | 0.196 | notch3 | -0.130 |
| vamp2 | 0.196 | vamp3 | -0.129 |
| ndrg4 | 0.196 | rpl23 | -0.129 |
| sncgb | 0.194 | rplp2l | -0.129 |
| rtn1a | 0.194 | si:ch73-281n10.2 | -0.129 |
| nsg2 | 0.192 | COX7A2 (1 of many) | -0.129 |
| map1aa | 0.189 | her12 | -0.128 |
| myt1b | 0.189 | tspan7 | -0.128 |
| eno2 | 0.188 | zgc:56493 | -0.127 |
| atp1b2a | 0.185 | gfap | -0.127 |
| kcnc3a | 0.185 | rpl39 | -0.127 |
| cplx2l | 0.185 | rps7 | -0.126 |
| aplp1 | 0.181 | btg2 | -0.125 |
| zgc:65894 | 0.178 | rps20 | -0.125 |
| csdc2a | 0.177 | rps15a | -0.123 |
| calm1a | 0.177 | msi1 | -0.122 |
| tuba2 | 0.176 | rpl7 | -0.122 |
| stmn4l | 0.176 | rpl10a | -0.119 |
| pacsin1a | 0.175 | pcna | -0.118 |