"filamin C, gamma b (actin binding protein 280)"
ZFIN 
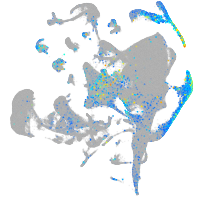
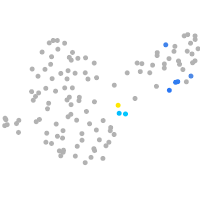
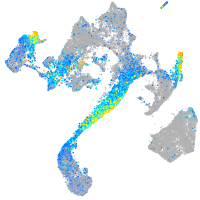
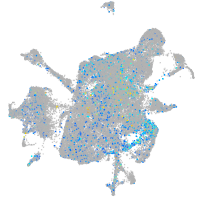
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX465205.2 | 0.238 | gstt1a | -0.067 |
| CU570689.1 | 0.234 | gapdh | -0.066 |
| zgc:162941 | 0.215 | prdx2 | -0.065 |
| prok1 | 0.210 | apoa4b.1 | -0.064 |
| si:ch211-203k16.3 | 0.204 | eno3 | -0.063 |
| XLOC-029278 | 0.201 | apoc2 | -0.061 |
| BX649398.3 | 0.185 | pklr | -0.061 |
| ccdc33 | 0.182 | gstr | -0.060 |
| CT573366.1 | 0.171 | sdr16c5b | -0.060 |
| XLOC-026848 | 0.166 | scp2a | -0.060 |
| si:ch73-190f9.4 | 0.164 | gcshb | -0.057 |
| si:dkey-88n24.7 | 0.164 | haao | -0.057 |
| loxl3a | 0.154 | tdo2a | -0.057 |
| tph1a | 0.149 | sult2st2 | -0.057 |
| LOC110438602 | 0.149 | rdh1 | -0.057 |
| LOC103910665 | 0.149 | srd5a2a | -0.056 |
| smtna | 0.147 | apoa1b | -0.056 |
| vgll2a | 0.146 | ugt1a7 | -0.055 |
| LOC103911081 | 0.146 | dhrs9 | -0.055 |
| CU302200.1 | 0.146 | upb1 | -0.055 |
| lygl2 | 0.145 | cox7a1 | -0.054 |
| si:ch73-265h17.2 | 0.138 | acmsd | -0.053 |
| CU682640.1 | 0.138 | gpx4a | -0.053 |
| fgf16 | 0.134 | aldh6a1 | -0.053 |
| RF00609 | 0.133 | sod1 | -0.053 |
| whrnb | 0.132 | dpydb | -0.053 |
| hapln4 | 0.130 | mdh1aa | -0.052 |
| mir133c | 0.129 | cat | -0.052 |
| CABZ01034691.1 | 0.129 | cox6b1 | -0.052 |
| p4htm | 0.129 | lgals2b | -0.052 |
| FQ311920.1 | 0.127 | fabp10a | -0.051 |
| myoc | 0.126 | gamt | -0.051 |
| si:rp71-77l1.1 | 0.124 | ftcd | -0.051 |
| LOC100001908 | 0.122 | aldh1l1 | -0.051 |
| myog | 0.118 | rgn | -0.051 |