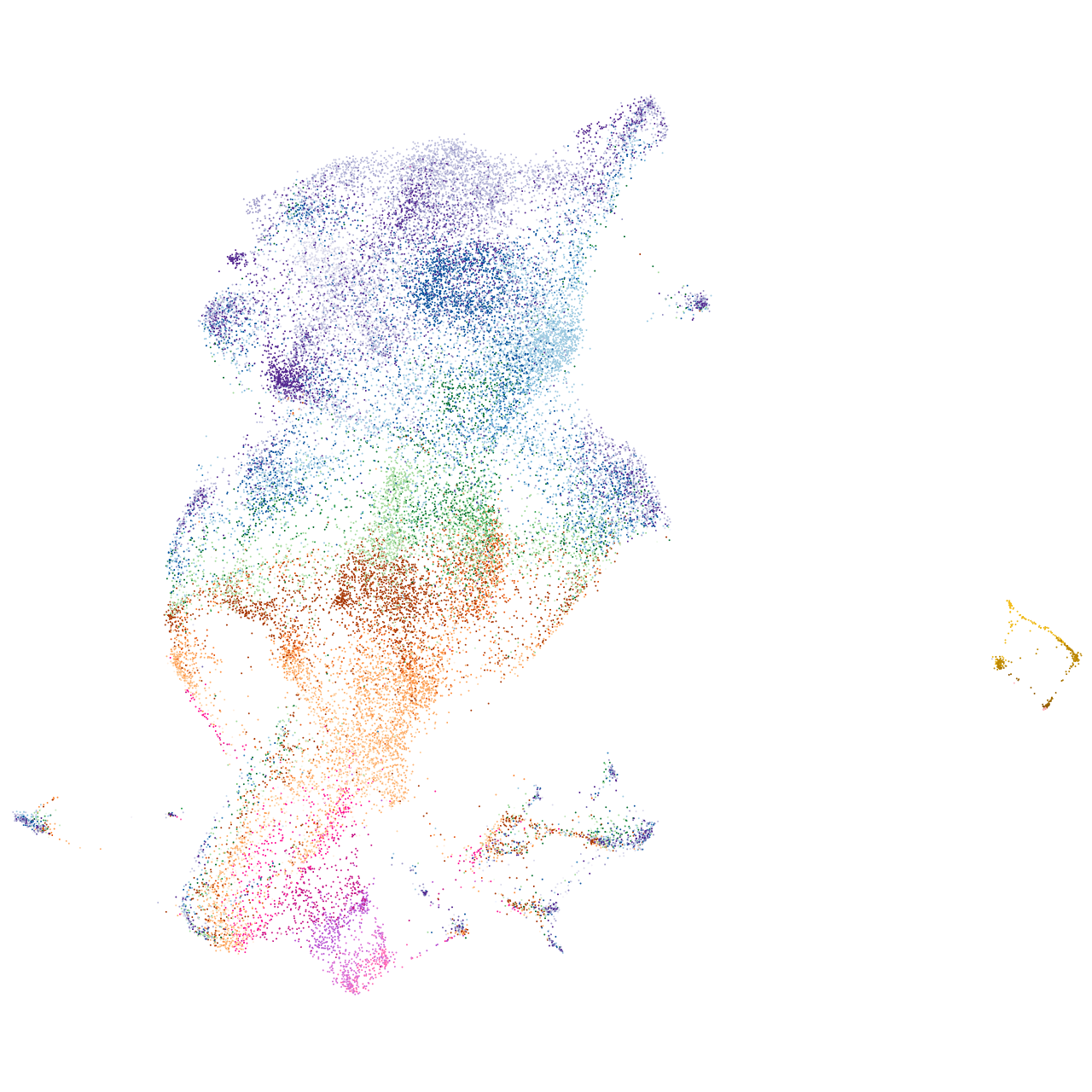FKBP prolyl isomerase 1Ab
ZFIN 
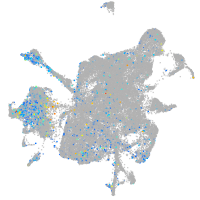
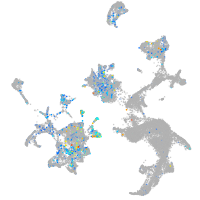
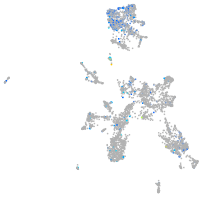
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb1b | 0.166 | cyt1 | -0.076 |
| si:ch211-133n4.4 | 0.165 | krt4 | -0.065 |
| myt1a | 0.163 | krt5 | -0.061 |
| scrt2 | 0.161 | si:ch211-195b11.3 | -0.061 |
| myt1b | 0.156 | wu:fb18f06 | -0.048 |
| gpm6aa | 0.154 | scel | -0.047 |
| cadm3 | 0.153 | cfl1l | -0.046 |
| ckbb | 0.152 | si:ch73-52f15.5 | -0.046 |
| gpm6ab | 0.152 | krt17 | -0.046 |
| hmgb3a | 0.151 | zgc:175088 | -0.046 |
| nova2 | 0.150 | anxa1a | -0.044 |
| si:ch211-137a8.4 | 0.150 | mgst1.2 | -0.044 |
| tuba1a | 0.150 | ckap4 | -0.043 |
| marcksl1b | 0.148 | agr1 | -0.042 |
| LOC100330246 | 0.148 | selenow1 | -0.042 |
| tubb5 | 0.148 | cyt1l | -0.041 |
| marcksl1a | 0.146 | slc38a5b | -0.041 |
| insm1a | 0.146 | icn2 | -0.041 |
| rtn1a | 0.145 | zgc:162730 | -0.040 |
| crx | 0.145 | eef1da | -0.040 |
| neurod4 | 0.144 | anxa1b | -0.037 |
| si:ch211-222l21.1 | 0.142 | krtt1c19e | -0.037 |
| stmn1b | 0.141 | s100v2 | -0.037 |
| nkain1 | 0.139 | si:ch211-157c3.4 | -0.036 |
| ncam1a | 0.138 | glo1 | -0.036 |
| mdkb | 0.138 | zgc:193505 | -0.035 |
| CU634008.1 | 0.137 | eevs | -0.035 |
| epb41a | 0.137 | abcb5 | -0.034 |
| tuba1c | 0.135 | si:dkey-16p21.8 | -0.034 |
| marcksb | 0.133 | pycard | -0.033 |
| thsd7aa | 0.132 | si:ch211-105c13.3 | -0.033 |
| dbn1 | 0.132 | mpc1 | -0.032 |
| vamp2 | 0.132 | tcima | -0.032 |
| tp53inp2 | 0.131 | tdh | -0.032 |
| robo3 | 0.131 | mcl1a | -0.032 |