"FEV transcription factor, ETS family member"
ZFIN 
Other cell groups


all cells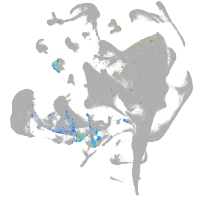 |
blastomeres |
PGCs |
endoderm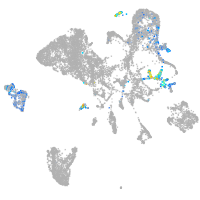 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 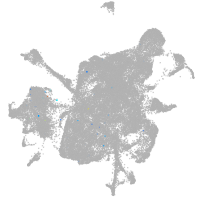 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia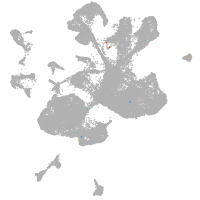 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tph2 | 0.557 | hmgb2a | -0.045 |
| hmx3b | 0.317 | rplp1 | -0.041 |
| ddc | 0.286 | hmgb2b | -0.039 |
| slc6a4a | 0.284 | rplp2l | -0.039 |
| txn | 0.248 | rps15a | -0.039 |
| slc18a2 | 0.236 | si:dkey-151g10.6 | -0.039 |
| lmx1ba | 0.223 | rps12 | -0.039 |
| lmx1al | 0.213 | rps28 | -0.037 |
| ucn3l | 0.197 | rpl10a | -0.036 |
| lmx1bb | 0.193 | rps20 | -0.036 |
| gch1 | 0.169 | rps7 | -0.035 |
| gata2a | 0.166 | rplp0 | -0.035 |
| pth1rb | 0.165 | rpsa | -0.034 |
| si:ch211-196h16.12 | 0.151 | rpl23 | -0.034 |
| LOC571123 | 0.145 | rpl11 | -0.034 |
| UNC13B | 0.142 | pcna | -0.033 |
| gata3 | 0.126 | rps9 | -0.033 |
| gfra1b | 0.119 | id1 | -0.033 |
| rpp25b | 0.117 | si:ch73-281n10.2 | -0.033 |
| mir375-2 | 0.116 | rpl7a | -0.033 |
| prdx1 | 0.109 | rps27a | -0.033 |
| penka | 0.104 | ran | -0.032 |
| calb2a | 0.102 | mdka | -0.032 |
| shox2 | 0.097 | rps4x | -0.032 |
| htr1d | 0.092 | rpl29 | -0.032 |
| oprl1 | 0.092 | rps2 | -0.032 |
| qdpra | 0.089 | rps5 | -0.031 |
| slc17a8 | 0.086 | rpl12 | -0.031 |
| BX664625.1 | 0.083 | rpl35a | -0.031 |
| cxcr5 | 0.078 | rpl35 | -0.031 |
| cpne7 | 0.078 | rpl36a | -0.031 |
| cryba2b | 0.075 | eef1a1l1 | -0.031 |
| cacng5a | 0.072 | rps19 | -0.031 |
| CABZ01056052.1 | 0.071 | rpl7 | -0.031 |
| AL845419.1 | 0.070 | h2afvb | -0.031 |




