ferredoxin 1
ZFIN 
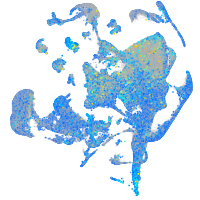
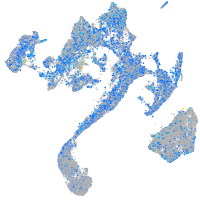
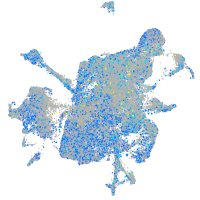
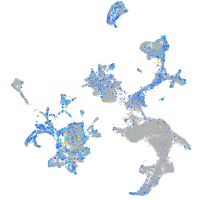
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fabp7a | 0.146 | elavl3 | -0.133 |
| atp1b4 | 0.137 | myt1b | -0.114 |
| slc3a2a | 0.134 | ptmab | -0.103 |
| id1 | 0.128 | stmn1b | -0.100 |
| si:ch1073-303k11.2 | 0.127 | tmsb | -0.097 |
| hepacama | 0.126 | onecut1 | -0.090 |
| ppap2d | 0.126 | elavl4 | -0.090 |
| si:ch211-66e2.5 | 0.125 | si:ch211-222l21.1 | -0.087 |
| cspg5b | 0.122 | stxbp1a | -0.086 |
| cd63 | 0.122 | zfhx3 | -0.086 |
| atp1a1b | 0.122 | zc4h2 | -0.086 |
| slc6a11b | 0.121 | gng3 | -0.086 |
| sept8b | 0.120 | tubb5 | -0.085 |
| GCA | 0.120 | stx1b | -0.085 |
| efhd1 | 0.119 | LOC100537384 | -0.085 |
| sdcbp2 | 0.119 | sncb | -0.085 |
| glula | 0.119 | si:ch73-386h18.1 | -0.085 |
| spry2 | 0.119 | gng2 | -0.084 |
| si:ch211-286b5.5 | 0.119 | pik3r3b | -0.083 |
| anxa13 | 0.117 | nsg2 | -0.083 |
| gnai2a | 0.117 | myt1a | -0.083 |
| vamp3 | 0.115 | rtn1a | -0.083 |
| psph | 0.115 | onecut2 | -0.082 |
| cx43 | 0.115 | ptmaa | -0.081 |
| eno1b | 0.115 | syt2a | -0.081 |
| si:dkey-7j14.6 | 0.115 | snap25a | -0.081 |
| ddt | 0.114 | gap43 | -0.080 |
| msi1 | 0.114 | rtn1b | -0.080 |
| sept10 | 0.114 | jagn1a | -0.078 |
| psat1 | 0.114 | vamp2 | -0.078 |
| gpr37l1b | 0.113 | golga7ba | -0.077 |
| nck1a | 0.113 | cplx2 | -0.077 |
| slc4a4a | 0.113 | rbfox3a | -0.076 |
| slc1a2b | 0.112 | scrt2 | -0.076 |
| endouc | 0.111 | rbfox1 | -0.075 |