F-box protein 5
ZFIN 
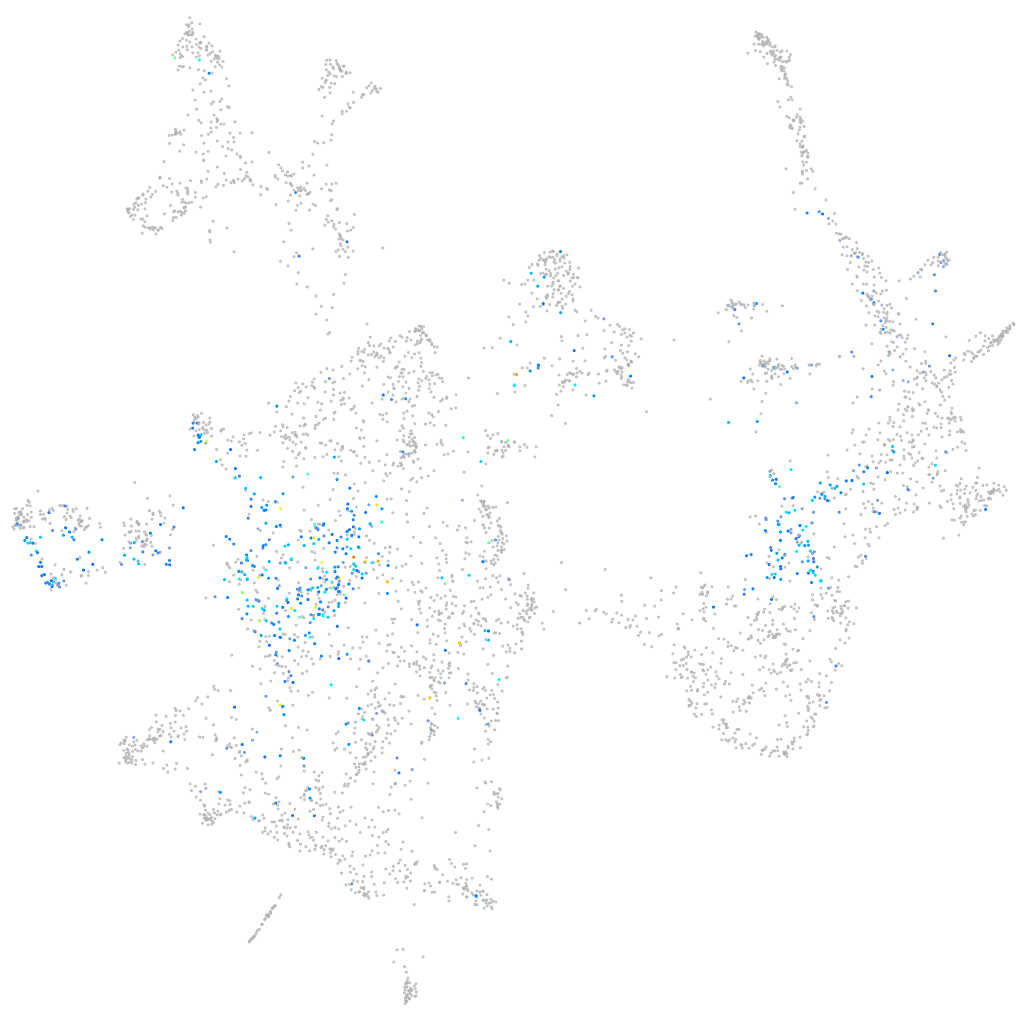
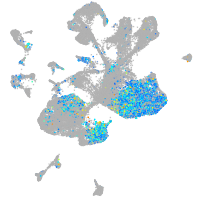
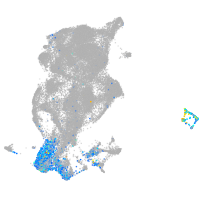
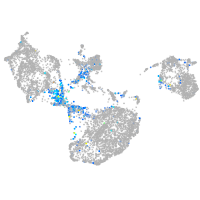
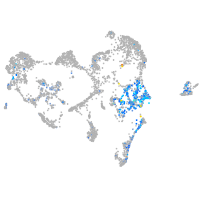
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cks1b | 0.547 | tob1b | -0.168 |
| mibp | 0.519 | nupr1a | -0.144 |
| aurkb | 0.519 | calm1a | -0.144 |
| rrm2 | 0.511 | gabarapb | -0.141 |
| ccna2 | 0.510 | ccni | -0.137 |
| zgc:165555.3 | 0.508 | tmem59 | -0.136 |
| mki67 | 0.505 | b2ml | -0.134 |
| mad2l1 | 0.503 | gapdhs | -0.134 |
| zgc:110216 | 0.494 | atp2b1a | -0.130 |
| cdk1 | 0.491 | pvalb8 | -0.130 |
| si:dkey-261m9.17 | 0.486 | eno1a | -0.128 |
| cdca5 | 0.485 | h1f0 | -0.127 |
| zgc:110540 | 0.478 | pvalb1 | -0.127 |
| dut | 0.468 | gabarapl2 | -0.127 |
| rrm1 | 0.467 | rnasekb | -0.125 |
| zgc:110425 | 0.466 | calml4a | -0.124 |
| stmn1a | 0.462 | atp5if1b | -0.124 |
| hist1h4l | 0.461 | otofb | -0.123 |
| dek | 0.457 | nptna | -0.122 |
| si:ch211-113a14.18 | 0.455 | gabarapa | -0.121 |
| top2a | 0.453 | si:dkey-16p21.8 | -0.120 |
| si:dkey-6i22.5 | 0.438 | tspan13b | -0.120 |
| CABZ01005379.1 | 0.437 | atp6v1e1b | -0.120 |
| pcna | 0.436 | calm1b | -0.119 |
| dhfr | 0.435 | gpx2 | -0.118 |
| tpx2 | 0.430 | pkig | -0.118 |
| tubb2b | 0.429 | ube2h | -0.117 |
| ncapd2 | 0.428 | selenow1 | -0.117 |
| esco2 | 0.427 | atp1a3b | -0.116 |
| ncapg | 0.424 | map1lc3a | -0.115 |
| chaf1a | 0.422 | twf2b | -0.115 |
| si:dkey-108k21.10 | 0.419 | capgb | -0.115 |
| smc2 | 0.415 | cd164l2 | -0.114 |
| smc4 | 0.415 | osbpl1a | -0.114 |
| lbr | 0.414 | socs3a | -0.113 |