"family with sequence similarity 57, member Ba [Source:ZFIN;Acc:ZDB-GENE-030131-2046]"
ZFIN 
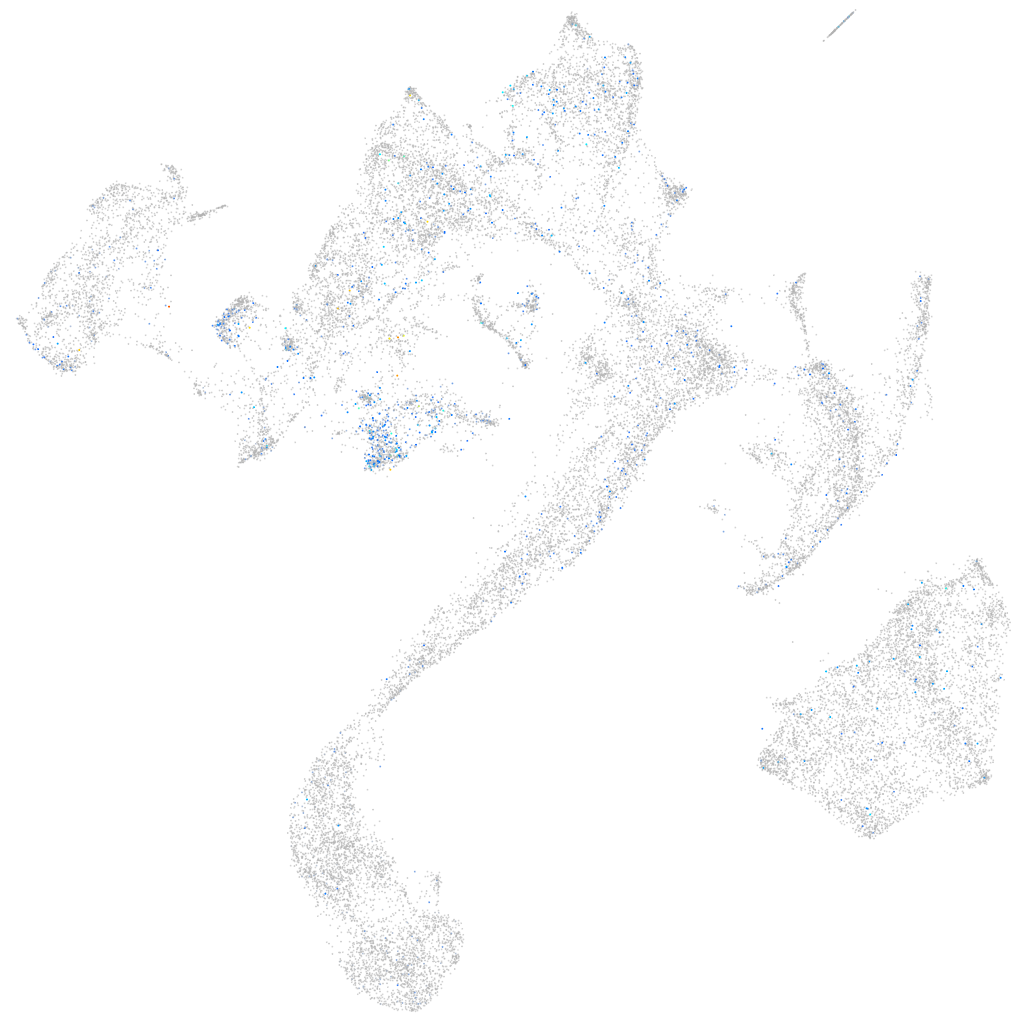
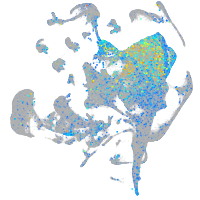
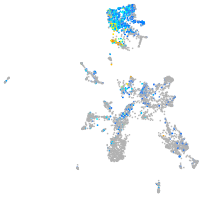
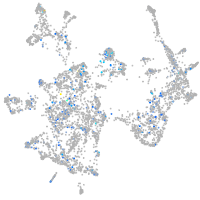
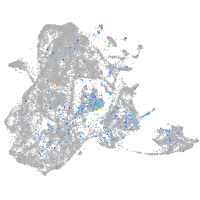
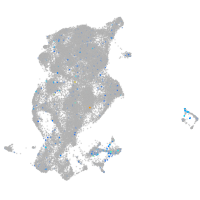
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpm6aa | 0.159 | hspb1 | -0.057 |
| stmn1b | 0.148 | apoc1 | -0.047 |
| elavl3 | 0.147 | tuba8l2 | -0.045 |
| fez1 | 0.134 | apoeb | -0.044 |
| tuba1c | 0.127 | tpma | -0.041 |
| ptmaa | 0.124 | tmod4 | -0.041 |
| epb41a | 0.122 | sp5l | -0.041 |
| myt1b | 0.121 | si:ch211-152c2.3 | -0.040 |
| rnasekb | 0.121 | polr3gla | -0.040 |
| ckbb | 0.120 | tnnc2 | -0.040 |
| rtn1b | 0.120 | cdx4 | -0.040 |
| cadm3 | 0.119 | myoz1b | -0.040 |
| vamp2 | 0.118 | XLOC-025819 | -0.040 |
| elavl4 | 0.116 | hhatla | -0.039 |
| snap25a | 0.115 | XLOC-006515 | -0.039 |
| nova2 | 0.115 | mybphb | -0.038 |
| fam117ba | 0.114 | ldb3b | -0.038 |
| sncb | 0.113 | CABZ01061524.1 | -0.038 |
| snap25b | 0.113 | CABZ01078594.1 | -0.038 |
| gng3 | 0.112 | tbx16 | -0.037 |
| thsd7aa | 0.112 | BX927258.1 | -0.037 |
| dpysl5a | 0.112 | prr33 | -0.037 |
| zgc:65894 | 0.111 | vox | -0.037 |
| stxbp1a | 0.111 | acta1b | -0.037 |
| ndrg4 | 0.111 | CR855257.1 | -0.037 |
| atp6v0cb | 0.111 | XLOC-005350 | -0.037 |
| nsg2 | 0.111 | mylpfb | -0.037 |
| scrt2 | 0.111 | LOC110440043 | -0.036 |
| scg2b | 0.109 | hacd1 | -0.036 |
| id4 | 0.109 | rtn2b | -0.036 |
| gpr85 | 0.108 | ddx18 | -0.036 |
| gpr27 | 0.107 | XLOC-001975 | -0.036 |
| syt1a | 0.107 | myom2a | -0.035 |
| aplp1 | 0.105 | si:ch73-367p23.2 | -0.035 |
| cspg5a | 0.105 | gar1 | -0.035 |