family with sequence similarity 234 member A
ZFIN 
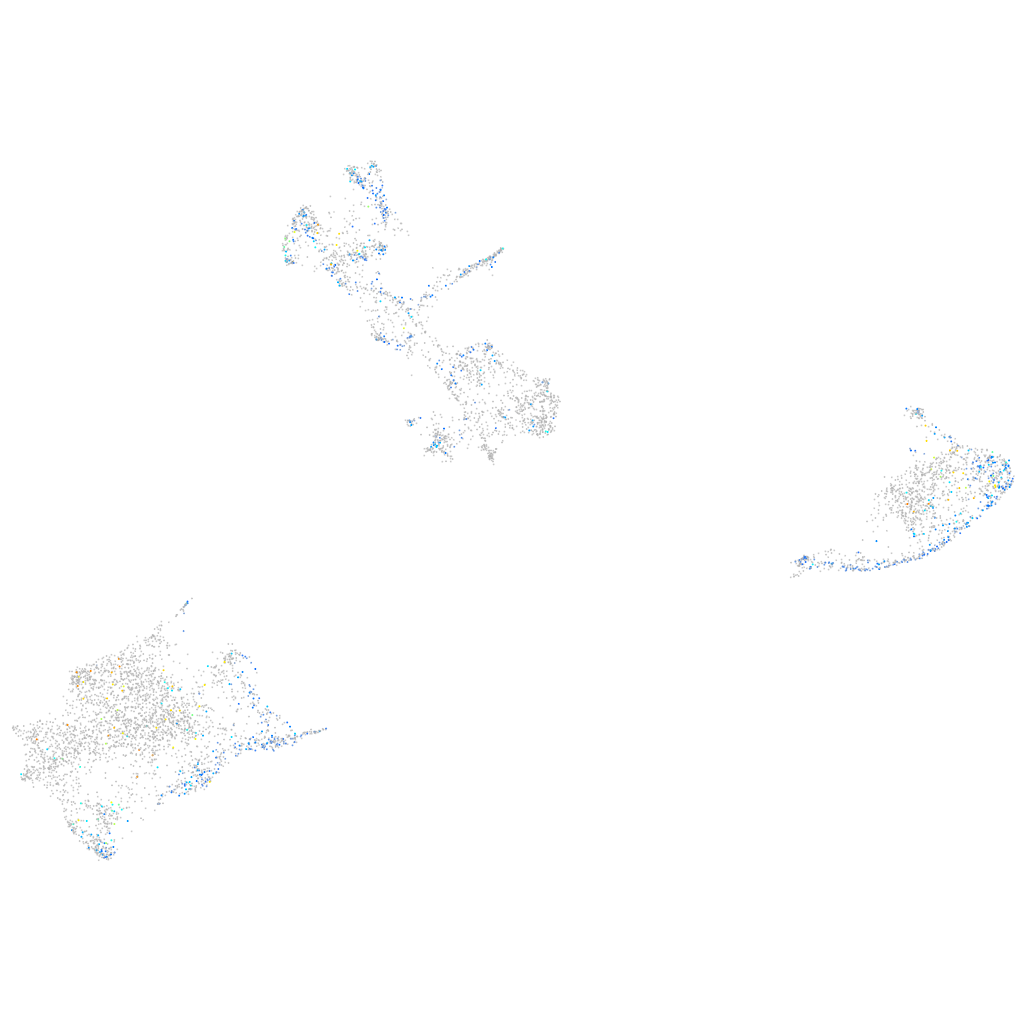
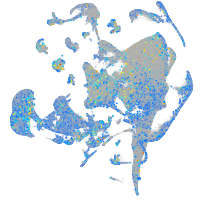
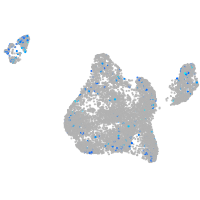
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.137 | CABZ01021592.1 | -0.078 |
| sptlc2a | 0.130 | si:dkey-251i10.2 | -0.069 |
| kita | 0.129 | gpm6aa | -0.061 |
| trpm1b | 0.128 | gch2 | -0.058 |
| BX571715.1 | 0.127 | nova2 | -0.058 |
| atp11a | 0.126 | ptmaa | -0.058 |
| si:zfos-943e10.1 | 0.125 | stmn1b | -0.056 |
| gadd45ba | 0.122 | si:ch211-222l21.1 | -0.056 |
| slc29a1a | 0.120 | epb41a | -0.056 |
| psap | 0.118 | si:ch73-1a9.3 | -0.055 |
| rnaset2 | 0.116 | elavl3 | -0.054 |
| adam10a | 0.115 | tmsb | -0.052 |
| slc45a2 | 0.114 | CU467822.1 | -0.050 |
| col4a1 | 0.113 | tuba1c | -0.050 |
| zgc:110239 | 0.112 | zc4h2 | -0.049 |
| adgrg2a | 0.111 | rplp0 | -0.049 |
| rhag | 0.111 | cadm3 | -0.048 |
| atp6v0ca | 0.110 | marcksl1a | -0.048 |
| bace2 | 0.108 | hmgb3a | -0.048 |
| ctsba | 0.108 | gng3 | -0.048 |
| sdc4 | 0.107 | elavl4 | -0.047 |
| tyr | 0.107 | gng2 | -0.046 |
| ctsla | 0.107 | sox11b | -0.046 |
| lyst | 0.106 | hmgb1b | -0.046 |
| ddit3 | 0.106 | sox11a | -0.045 |
| opn5 | 0.105 | hbae1.1 | -0.044 |
| cdh1 | 0.103 | mllt11 | -0.044 |
| lamb1b | 0.102 | tmeff1b | -0.044 |
| canx | 0.101 | actc1b | -0.043 |
| LOC103909099 | 0.101 | XLOC-003692 | -0.043 |
| si:dkey-56m15.5 | 0.100 | mir181b-3 | -0.042 |
| slc39a10 | 0.099 | sncb | -0.042 |
| dtnbp1a | 0.099 | CR759927.3 | -0.042 |
| col4a3bpb | 0.098 | tox | -0.041 |
| kank2 | 0.098 | her15.1 | -0.041 |