family with sequence similarity 189 member A2
ZFIN 
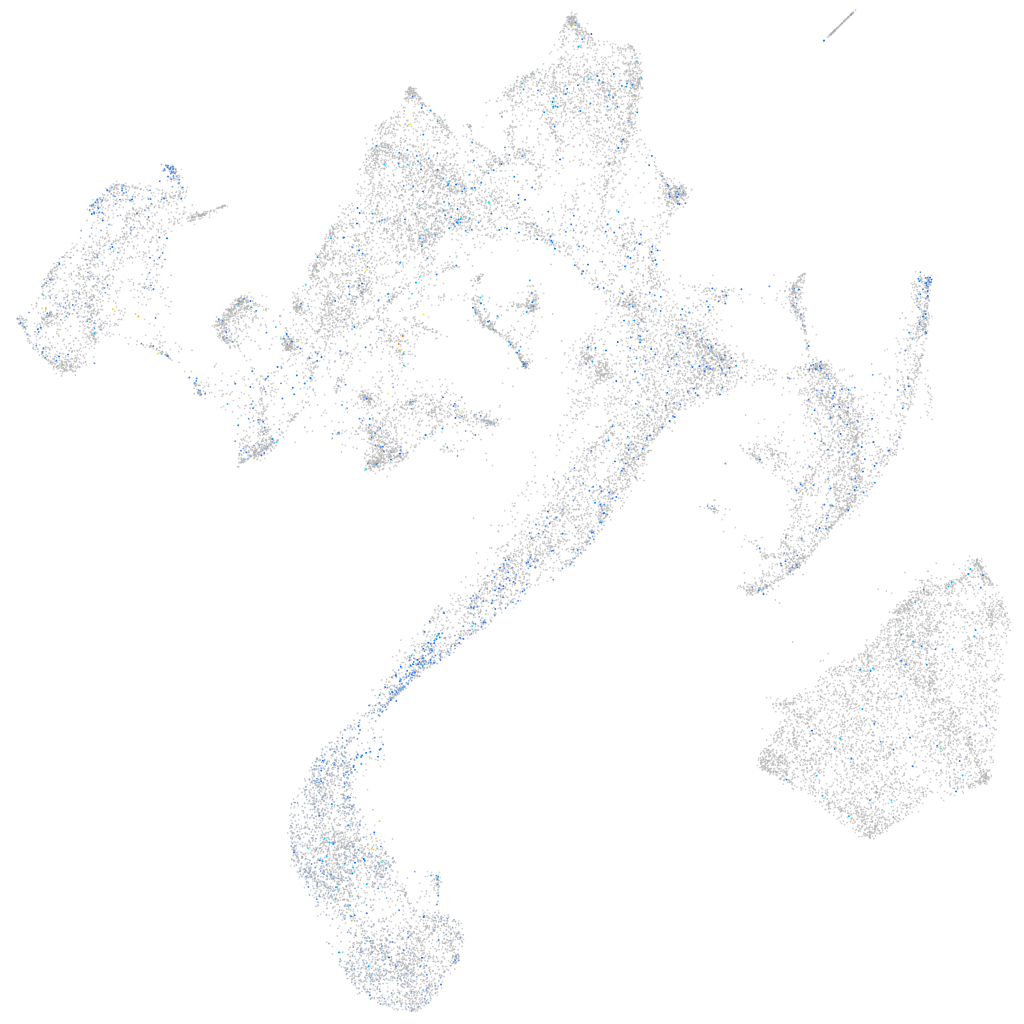
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01072309.1 | 0.143 | si:ch73-1a9.3 | -0.115 |
| trdn | 0.143 | ptmab | -0.112 |
| srl | 0.137 | hmgb2b | -0.110 |
| tmem38a | 0.136 | hmgb2a | -0.109 |
| cav3 | 0.136 | pabpc1a | -0.109 |
| ryr1b | 0.136 | hsp90ab1 | -0.103 |
| ttn.2 | 0.135 | si:ch211-222l21.1 | -0.101 |
| rtn2a | 0.133 | seta | -0.101 |
| neb | 0.132 | si:ch73-281n10.2 | -0.100 |
| ttn.1 | 0.131 | hnrnpabb | -0.100 |
| ldb3a | 0.131 | anp32b | -0.100 |
| atp2a1 | 0.130 | hnrnpaba | -0.099 |
| usp13 | 0.129 | hmga1a | -0.099 |
| mylpfa | 0.129 | tuba8l4 | -0.099 |
| acta1b | 0.127 | khdrbs1a | -0.098 |
| plecb | 0.127 | anp32a | -0.098 |
| cavin4b | 0.126 | cirbpa | -0.097 |
| si:ch211-266g18.10 | 0.126 | hmgn7 | -0.096 |
| hhatla | 0.125 | si:ch211-288g17.3 | -0.095 |
| desma | 0.125 | h3f3d | -0.095 |
| tgm2a | 0.125 | hmgn2 | -0.095 |
| fitm1 | 0.125 | hnrnpa0b | -0.094 |
| si:ch73-367p23.2 | 0.125 | nucks1a | -0.093 |
| nmrk2 | 0.125 | sumo3a | -0.092 |
| slc8a3 | 0.124 | hnrnpa1b | -0.091 |
| klhl31 | 0.124 | h2afvb | -0.090 |
| casq2 | 0.124 | syncrip | -0.090 |
| XLOC-040108 | 0.124 | setb | -0.089 |
| CABZ01078594.1 | 0.123 | fthl27 | -0.089 |
| tpma | 0.123 | cx43.4 | -0.089 |
| zgc:158296 | 0.123 | cirbpb | -0.088 |
| mybphb | 0.123 | ran | -0.088 |
| CABZ01072309.2 | 0.123 | cbx3a | -0.087 |
| CABZ01072245.1 | 0.122 | top1l | -0.087 |
| aldoab | 0.122 | hnrnpub | -0.086 |