FA core complex associated protein 24
ZFIN 
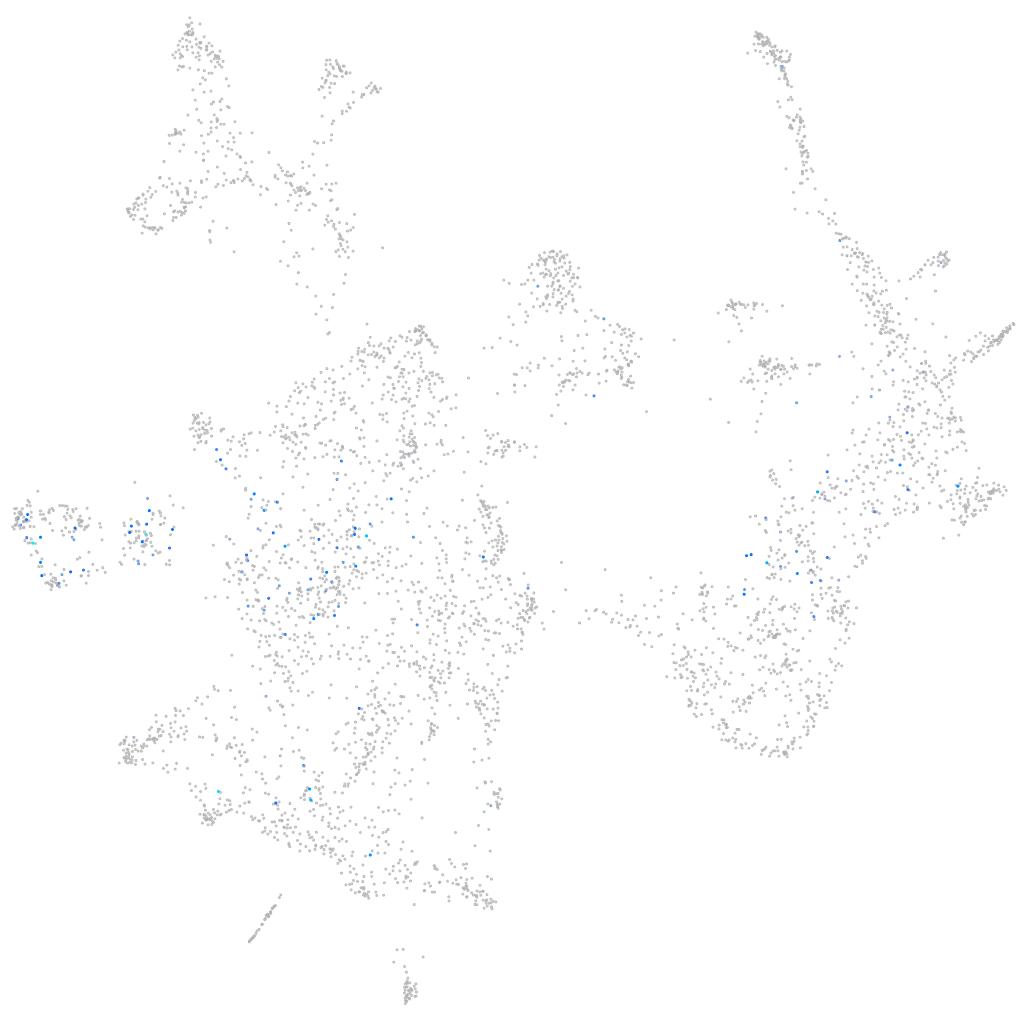
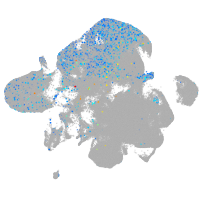
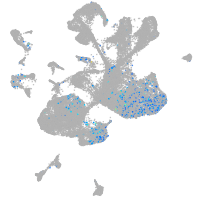
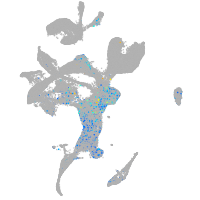
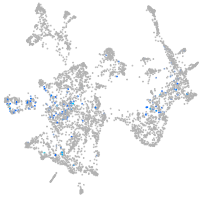
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cks1b | 0.251 | COX3 | -0.076 |
| zgc:110425 | 0.229 | tob1b | -0.075 |
| ccna2 | 0.228 | zgc:158463 | -0.073 |
| rrm1 | 0.228 | rplp2 | -0.071 |
| mki67 | 0.222 | atp2b1a | -0.067 |
| cdca5 | 0.215 | gabarapa | -0.064 |
| mad2l1 | 0.212 | si:dkey-16p21.8 | -0.064 |
| cdk1 | 0.208 | mt-co2 | -0.061 |
| znf1045 | 0.204 | b2ml | -0.061 |
| rrm2 | 0.202 | tpi1a | -0.060 |
| tpx2 | 0.201 | eno1a | -0.060 |
| si:dkey-6i22.5 | 0.195 | cpeb4b | -0.058 |
| pcna | 0.194 | otofb | -0.058 |
| CR855311.2 | 0.193 | atp6v1e1b | -0.058 |
| zgc:110216 | 0.189 | atp6v1g1 | -0.058 |
| dtymk | 0.189 | nptna | -0.058 |
| dut | 0.188 | calm1b | -0.057 |
| cenpe | 0.188 | tspan13b | -0.057 |
| hmgb2a | 0.187 | vamp2 | -0.057 |
| prim2 | 0.186 | atp1a3b | -0.056 |
| numa1 | 0.186 | igfbp5b | -0.056 |
| smc2 | 0.184 | calml4a | -0.056 |
| tubb2b | 0.184 | ptbp3 | -0.056 |
| plk1 | 0.184 | gpx2 | -0.056 |
| aurkb | 0.183 | cd164l2 | -0.056 |
| cdca8 | 0.183 | gabarapl2 | -0.056 |
| kif23 | 0.181 | pvalb9 | -0.056 |
| hist1h2a9 | 0.181 | rnasekb | -0.055 |
| mibp | 0.179 | gapdhs | -0.055 |
| zgc:165555.3 | 0.179 | nupr1a | -0.055 |
| ska1 | 0.179 | atp6v0cb | -0.055 |
| dek | 0.178 | kif1aa | -0.055 |
| suv39h1b | 0.177 | lrrc73 | -0.054 |
| tuba8l | 0.177 | atp5if1b | -0.054 |
| mus81 | 0.176 | gb:bc139872 | -0.054 |