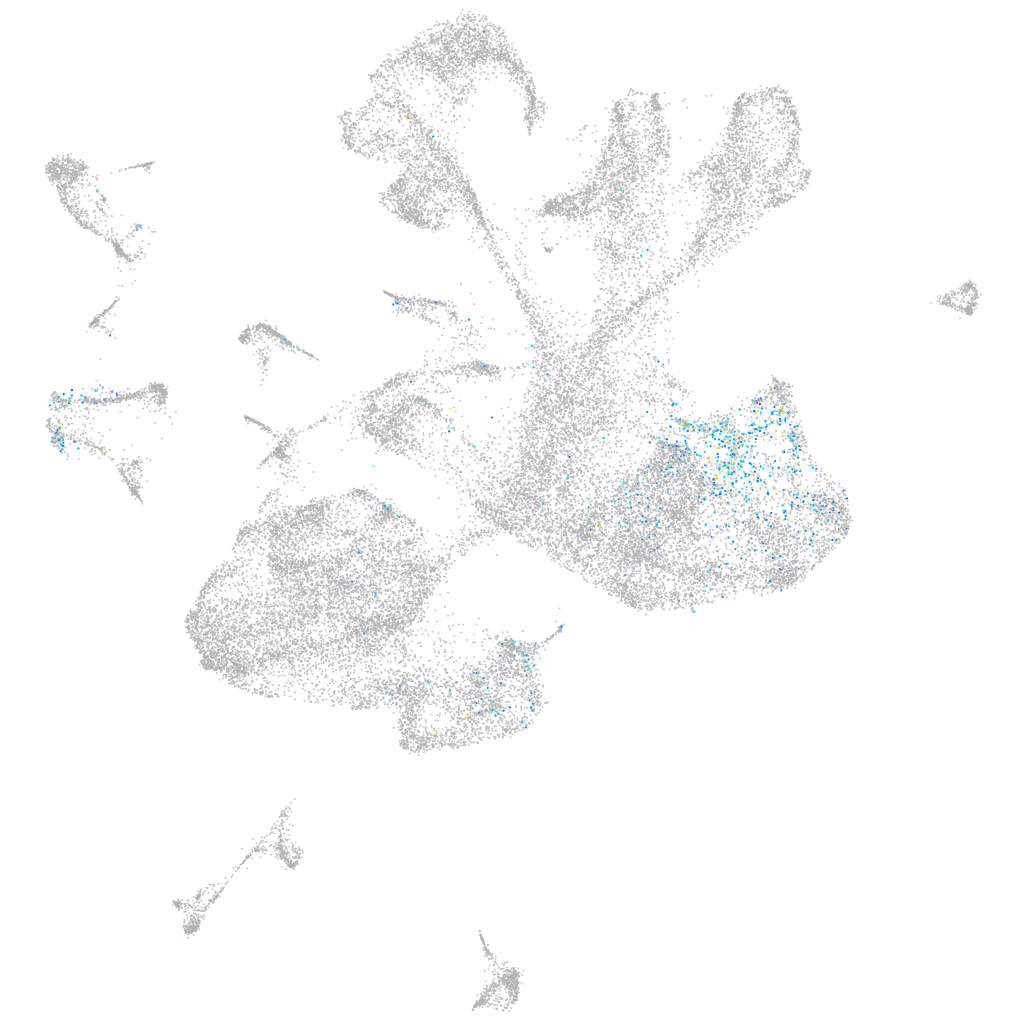
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.200 | ckbb | -0.095 |
| zgc:55461 | 0.199 | rtn1a | -0.081 |
| smkr1 | 0.188 | mdkb | -0.079 |
| foxj1a | 0.181 | rnasekb | -0.071 |
| si:ch211-163l21.7 | 0.173 | gpm6ab | -0.069 |
| nme5 | 0.167 | marcksl1b | -0.068 |
| dpcd | 0.165 | CU467822.1 | -0.068 |
| spag6 | 0.163 | stmn1b | -0.065 |
| ccdc173 | 0.158 | myt1b | -0.065 |
| efcab1 | 0.157 | elavl3 | -0.065 |
| rsph9 | 0.157 | slc1a2b | -0.064 |
| si:dkeyp-110a12.4 | 0.155 | tmsb | -0.064 |
| cfap52 | 0.153 | tmsb4x | -0.063 |
| ribc2 | 0.152 | COX3 | -0.063 |
| dnali1 | 0.152 | pvalb1 | -0.062 |
| cfap126 | 0.151 | ywhah | -0.062 |
| dnaaf4 | 0.149 | fez1 | -0.062 |
| cks1b | 0.146 | fabp7a | -0.059 |
| cdc20 | 0.146 | pvalb2 | -0.058 |
| mapk15 | 0.145 | dbn1 | -0.057 |
| ankrd45 | 0.145 | nova2 | -0.057 |
| gdf6b | 0.144 | dpysl3 | -0.056 |
| efcab2 | 0.143 | si:dkeyp-75h12.5 | -0.056 |
| ccnb1 | 0.143 | gng3 | -0.054 |
| enkur | 0.143 | atp6v1e1b | -0.054 |
| tex9 | 0.143 | atp6v0cb | -0.054 |
| aurkb | 0.143 | elavl4 | -0.054 |
| ttc25 | 0.142 | si:ch211-133n4.4 | -0.053 |
| mad2l1 | 0.142 | gpm6aa | -0.053 |
| ccdc151 | 0.141 | si:dkey-276j7.1 | -0.053 |
| si:dkeyp-69c1.9 | 0.141 | sncb | -0.052 |
| si:ch211-248e11.2 | 0.139 | CR383676.1 | -0.052 |
| dydc2 | 0.138 | vamp2 | -0.051 |
| spata4 | 0.138 | syt11a | -0.051 |
| LOC101884799 | 0.137 | atp6v1g1 | -0.051 |