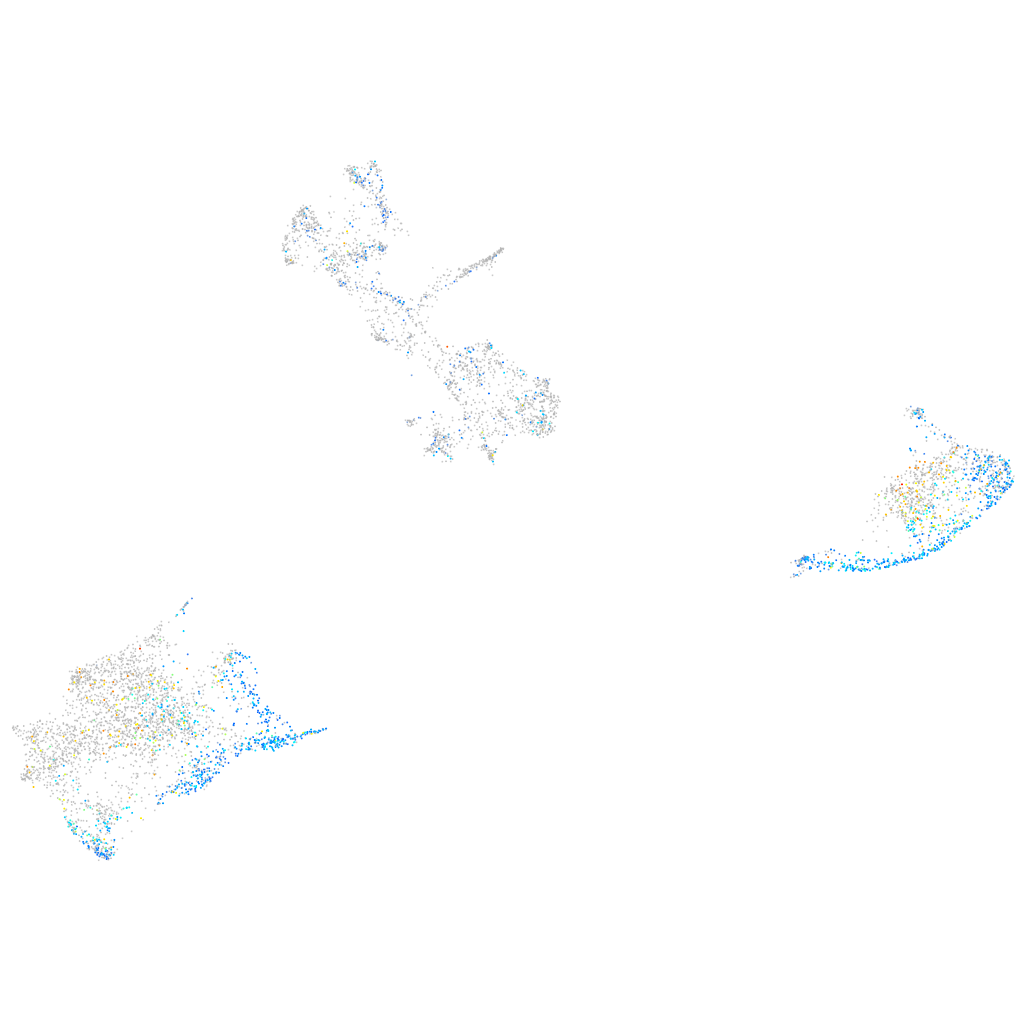
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mitfa | 0.293 | defbl1 | -0.153 |
| tspan36 | 0.279 | apoda.1 | -0.150 |
| tfap2e | 0.271 | mdkb | -0.142 |
| tmem243a | 0.256 | si:ch211-256m1.8 | -0.138 |
| tspan10 | 0.256 | ptmaa | -0.135 |
| tyr | 0.254 | gpnmb | -0.135 |
| tmem243b | 0.247 | lypc | -0.132 |
| slc3a2a | 0.245 | si:dkey-197i20.6 | -0.127 |
| qdpra | 0.244 | alx4a | -0.122 |
| rabl6b | 0.242 | si:ch73-89b15.3 | -0.119 |
| bace2 | 0.239 | gpm6aa | -0.118 |
| msx1b | 0.239 | unm-sa821 | -0.115 |
| C12orf75 | 0.239 | alx4b | -0.112 |
| syngr1a | 0.238 | cdh11 | -0.112 |
| pmela | 0.236 | tuba1c | -0.112 |
| rab38 | 0.235 | akap12a | -0.112 |
| pah | 0.234 | actc1b | -0.111 |
| slc37a2 | 0.232 | s100v2 | -0.108 |
| dct | 0.231 | nova2 | -0.108 |
| pcdh10a | 0.226 | alx1 | -0.107 |
| si:ch73-389b16.1 | 0.226 | prx | -0.106 |
| tyrp1a | 0.224 | pvalb1 | -0.106 |
| lamp1a | 0.222 | pvalb2 | -0.105 |
| tyrp1b | 0.221 | ponzr1 | -0.105 |
| zgc:91968 | 0.220 | hmgb1b | -0.102 |
| SPAG9 | 0.219 | sytl2b | -0.101 |
| pepd | 0.218 | stmn1b | -0.100 |
| pcbd1 | 0.216 | si:ch211-105c13.3 | -0.099 |
| xbp1 | 0.215 | slc25a36a | -0.099 |
| slc24a5 | 0.214 | hmgn2 | -0.097 |
| slc45a2 | 0.212 | si:ch73-1a9.3 | -0.096 |
| atp6v0ca | 0.211 | elavl3 | -0.096 |
| klf6a | 0.209 | sulf2b | -0.094 |
| naga | 0.209 | myof | -0.092 |
| degs1 | 0.209 | tcirg1a | -0.092 |