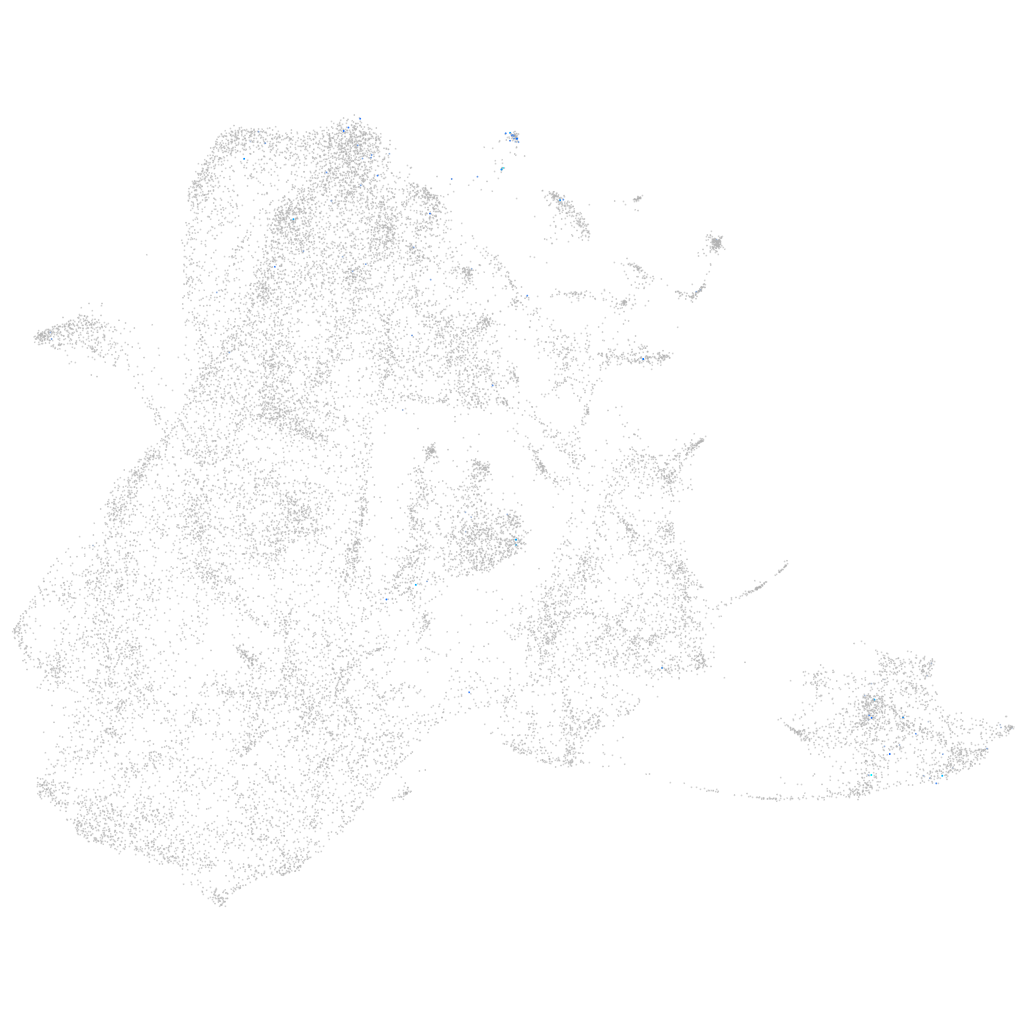
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye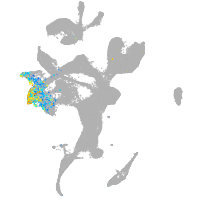 |
taste / olfactory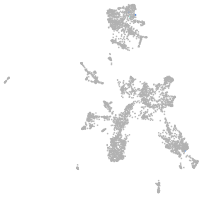 |
otic / lateral line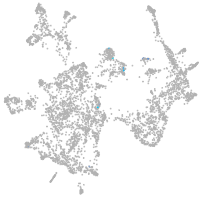 |
epidermis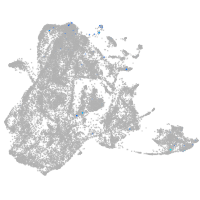 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-85k7.12 | 0.265 | tmsb4x | -0.033 |
| rbp4l | 0.208 | si:ch211-222l21.1 | -0.032 |
| lgals17 | 0.202 | tp63 | -0.032 |
| LOC100535225 | 0.201 | crtap | -0.032 |
| CT573282.1 | 0.201 | bcam | -0.032 |
| grk1b | 0.187 | fkbp9 | -0.031 |
| opn1lw2 | 0.186 | cldni | -0.031 |
| FO904966.2 | 0.179 | pak2a | -0.031 |
| panx2 | 0.171 | zgc:153675 | -0.031 |
| si:rp71-39b20.4 | 0.169 | tpm4a | -0.030 |
| LOC103910313 | 0.166 | cd151l | -0.030 |
| gnb3b | 0.165 | ywhaz | -0.029 |
| prph2b | 0.165 | fermt1 | -0.029 |
| grk7a | 0.165 | actb1 | -0.029 |
| rhol | 0.164 | col18a1a | -0.028 |
| gnat2 | 0.163 | myh9a | -0.028 |
| prph2a | 0.162 | kctd5a | -0.027 |
| rbp3 | 0.161 | dag1 | -0.027 |
| si:ch211-81a5.8 | 0.160 | fthl27 | -0.027 |
| pde6h | 0.157 | ssr4 | -0.027 |
| LOC103910500 | 0.157 | prdx4 | -0.027 |
| guk1b | 0.154 | tspan7 | -0.026 |
| slc25a3a | 0.148 | NC-002333.4 | -0.026 |
| zgc:162182 | 0.146 | fgfbp1b | -0.026 |
| pde6ha | 0.145 | snrpg | -0.026 |
| si:dkey-17e16.15 | 0.144 | fkbp14 | -0.026 |
| ckmt2a | 0.143 | p4ha1b | -0.026 |
| rcvrna | 0.143 | zgc:92380 | -0.025 |
| pdcb | 0.142 | id3 | -0.025 |
| si:ch1073-303d10.1 | 0.141 | plod3 | -0.025 |
| gngt2b | 0.141 | alcama | -0.025 |
| sagb | 0.139 | p3h1 | -0.025 |
| rcvrn2 | 0.137 | fkbp7 | -0.025 |
| gip | 0.137 | map2k6 | -0.025 |
| gngt1 | 0.135 | flot2a | -0.025 |