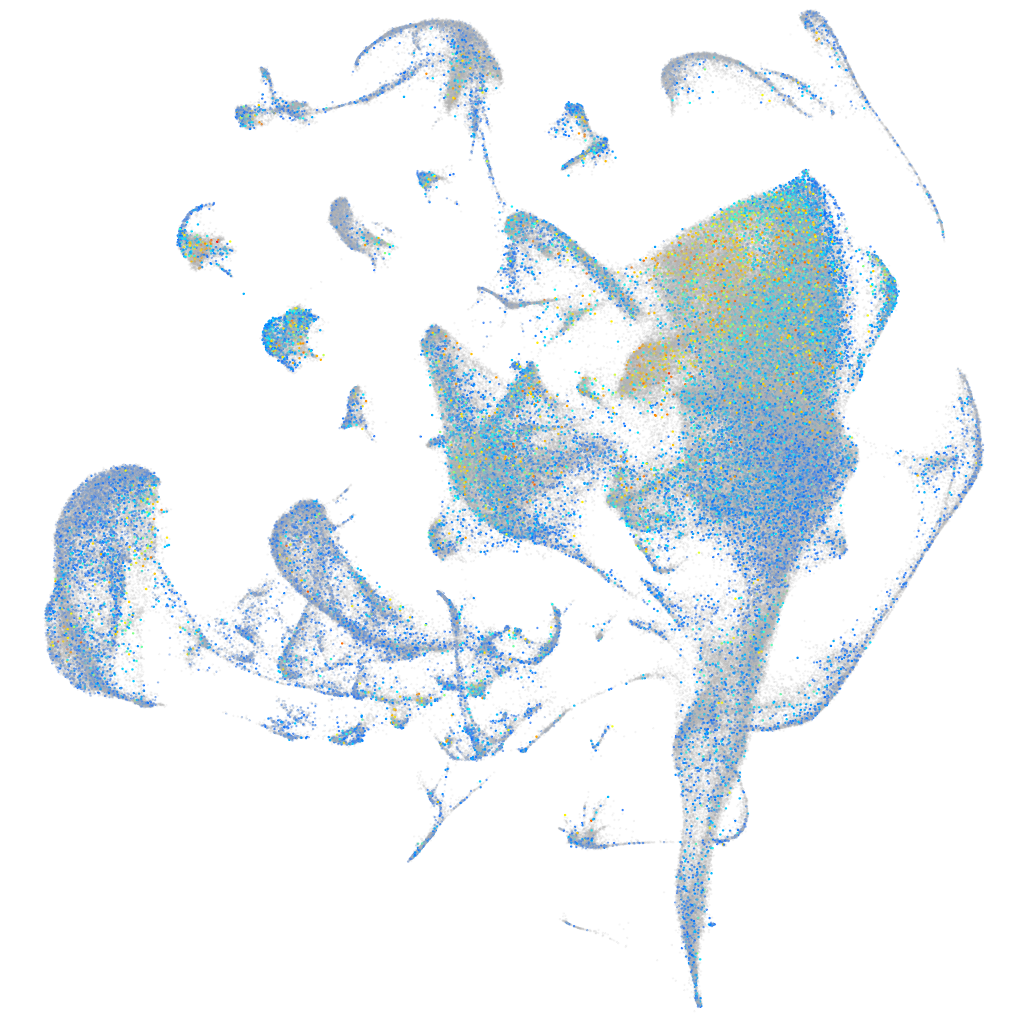exocyst complex component 7
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.068 | hspb1 | -0.048 |
| atp6v1g1 | 0.065 | npm1a | -0.044 |
| gabarapb | 0.064 | dkc1 | -0.042 |
| calm1a | 0.063 | nop58 | -0.041 |
| ywhah | 0.062 | fbl | -0.040 |
| gng3 | 0.061 | apoc1 | -0.039 |
| gnb1a | 0.060 | si:ch211-152c2.3 | -0.038 |
| stmn1b | 0.059 | lig1 | -0.037 |
| calm2a | 0.058 | polr3gla | -0.037 |
| mllt11 | 0.058 | stm | -0.037 |
| sncb | 0.058 | nop2 | -0.036 |
| tuba1c | 0.058 | zmp:0000000624 | -0.036 |
| zgc:65894 | 0.058 | ddx18 | -0.035 |
| ccni | 0.056 | gar1 | -0.035 |
| gng2 | 0.056 | pou5f3 | -0.035 |
| jpt1b | 0.056 | snu13b | -0.035 |
| gpm6ab | 0.055 | nop56 | -0.034 |
| hist2h2l | 0.055 | apoeb | -0.033 |
| rtn1b | 0.055 | pprc1 | -0.033 |
| stmn2a | 0.055 | si:dkey-66i24.9 | -0.033 |
| dpysl2b | 0.055 | banf1 | -0.032 |
| fez1 | 0.054 | bms1 | -0.032 |
| gabarapl2 | 0.054 | chaf1a | -0.032 |
| gapdhs | 0.054 | rrp15 | -0.032 |
| id4 | 0.054 | s100a1 | -0.032 |
| rnasekb | 0.054 | vox | -0.032 |
| vamp2 | 0.054 | BX927258.1 | -0.031 |
| si:ch1073-429i10.3.1 | 0.054 | gnl3 | -0.031 |
| cdc42 | 0.053 | ing5b | -0.031 |
| elavl3 | 0.053 | nop10 | -0.031 |
| gdi1 | 0.053 | pcna | -0.031 |
| atp6v1h | 0.052 | si:dkey-239h2.3 | -0.031 |
| ckbb | 0.052 | tbx16 | -0.031 |
| gabarapa | 0.052 | zgc:56699 | -0.031 |
| gap43 | 0.052 | anp32b | -0.030 |