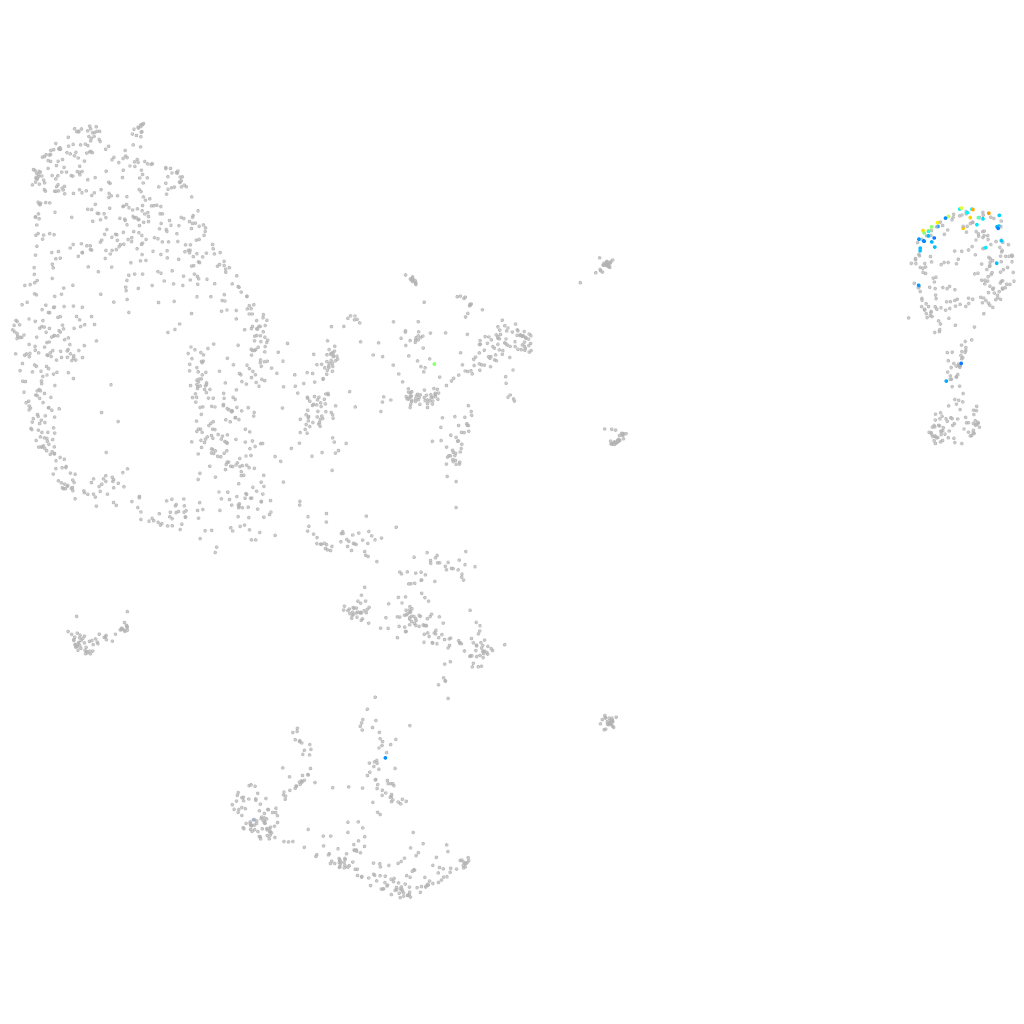
even-skipped-like1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 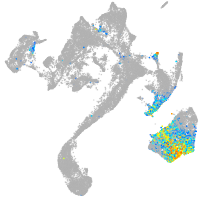 |
mural cells / non-skeletal muscle  |
mesenchyme 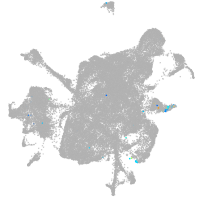 |
hematopoietic / vasculature  |
pronephros 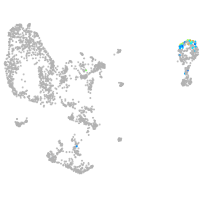 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ved | 0.568 | rpl37 | -0.286 |
| cdx1a | 0.455 | rps10 | -0.283 |
| tbx16 | 0.431 | zgc:114188 | -0.269 |
| szl | 0.423 | rps17 | -0.244 |
| vox | 0.413 | atp5f1b | -0.242 |
| cdx4 | 0.399 | atp1b1a | -0.239 |
| bambia | 0.369 | atp5l | -0.234 |
| apoc1 | 0.368 | cox8a | -0.226 |
| vent | 0.360 | zgc:56493 | -0.213 |
| XLOC-015209 | 0.352 | atp5mc3b | -0.212 |
| apoeb | 0.349 | atp5fa1 | -0.212 |
| BX927327.1 | 0.330 | atp5mc1 | -0.198 |
| tbx16l | 0.305 | atp5pb | -0.197 |
| aplnra | 0.287 | mt-nd1 | -0.196 |
| anp32e | 0.286 | atp5pf | -0.195 |
| blf | 0.286 | slc25a5 | -0.193 |
| cx43.4 | 0.286 | suclg1 | -0.191 |
| znfl2a | 0.285 | cox6a1 | -0.187 |
| si:ch211-106e7.2 | 0.281 | atp5f1e | -0.186 |
| wnt5b | 0.280 | cox7a2a | -0.186 |
| hspb1 | 0.276 | cdh17 | -0.186 |
| ggact.2 | 0.269 | atp5f1c | -0.185 |
| si:dkey-261j4.5 | 0.264 | idh2 | -0.184 |
| nop2 | 0.263 | vdac3 | -0.184 |
| srsf3a | 0.262 | vdac2 | -0.184 |
| dhx16 | 0.258 | mdh2 | -0.183 |
| msx2b | 0.258 | romo1 | -0.183 |
| crabp2b | 0.257 | hnrnpa0l | -0.182 |
| srrm1 | 0.257 | atp5meb | -0.181 |
| hsd3b2 | 0.256 | cox5aa | -0.180 |
| tmem136a | 0.256 | slc25a3b | -0.178 |
| rrp1 | 0.256 | cox4i1 | -0.177 |
| akap12b | 0.255 | ndufa4l | -0.176 |
| nop58 | 0.254 | ndrg1a | -0.173 |
| crygs4 | 0.253 | atp5if1b | -0.170 |