ETS variant transcription factor 5b
ZFIN 
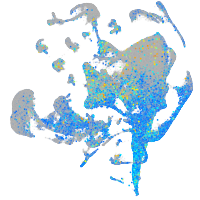
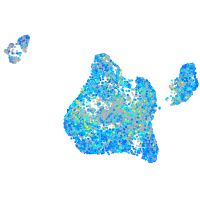
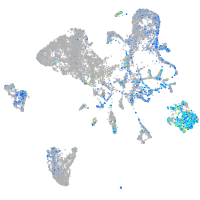
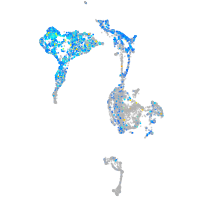
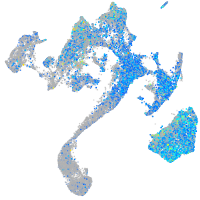
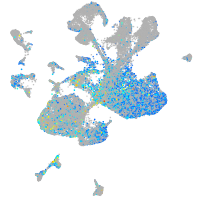
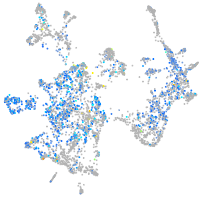
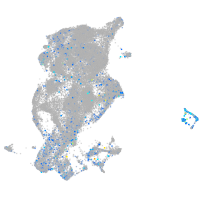
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cpne2 | 0.333 | si:dkey-36i7.3 | -0.182 |
| mfsd4ab | 0.274 | tmsb4x | -0.174 |
| kcnj13 | 0.261 | soul2 | -0.155 |
| slc26a6 | 0.252 | si:ch73-359m17.9 | -0.152 |
| aspa | 0.252 | atp1a1a.3 | -0.150 |
| etv5a | 0.244 | agr2 | -0.145 |
| slc5a8l | 0.236 | ppp1r1b | -0.144 |
| trpm7 | 0.230 | krt4 | -0.142 |
| st3gal7 | 0.229 | tbx2b | -0.141 |
| ace | 0.222 | bsnd | -0.140 |
| satb2 | 0.217 | ckmt1 | -0.139 |
| hsd17b3 | 0.216 | rnf183 | -0.138 |
| slc26a1 | 0.214 | zgc:158423 | -0.136 |
| atp2b2 | 0.213 | hbae3 | -0.136 |
| ptp4a2b | 0.210 | tuba1c | -0.135 |
| si:ch211-224l10.4 | 0.210 | ctgfa | -0.135 |
| epas1a | 0.207 | fabp3 | -0.135 |
| pdzd3a | 0.205 | trim35-12 | -0.133 |
| cers2a | 0.204 | dbn1 | -0.130 |
| eps8l3b | 0.204 | pfn1 | -0.129 |
| pvalb9 | 0.200 | slc7a8a | -0.127 |
| si:dkey-197j19.5 | 0.197 | gnao1b | -0.126 |
| wnk1a | 0.197 | elavl3 | -0.125 |
| arg1 | 0.196 | stmn1b | -0.125 |
| clic5b | 0.196 | actb1 | -0.124 |
| ahcyl2 | 0.195 | gdpd3a | -0.123 |
| zgc:152863 | 0.195 | clcnk | -0.123 |
| etnk1 | 0.194 | ahnak | -0.122 |
| atp11c | 0.192 | mal2 | -0.121 |
| zgc:162989 | 0.192 | AL954322.2 | -0.121 |
| slc6a6b | 0.190 | celf2 | -0.121 |
| serpinf2b | 0.189 | sesn1 | -0.120 |
| pdzk1ip1 | 0.188 | penkb | -0.120 |
| si:ch73-352p4.8 | 0.188 | slc12a3 | -0.120 |
| slc9a3.1 | 0.186 | rtn1a | -0.119 |