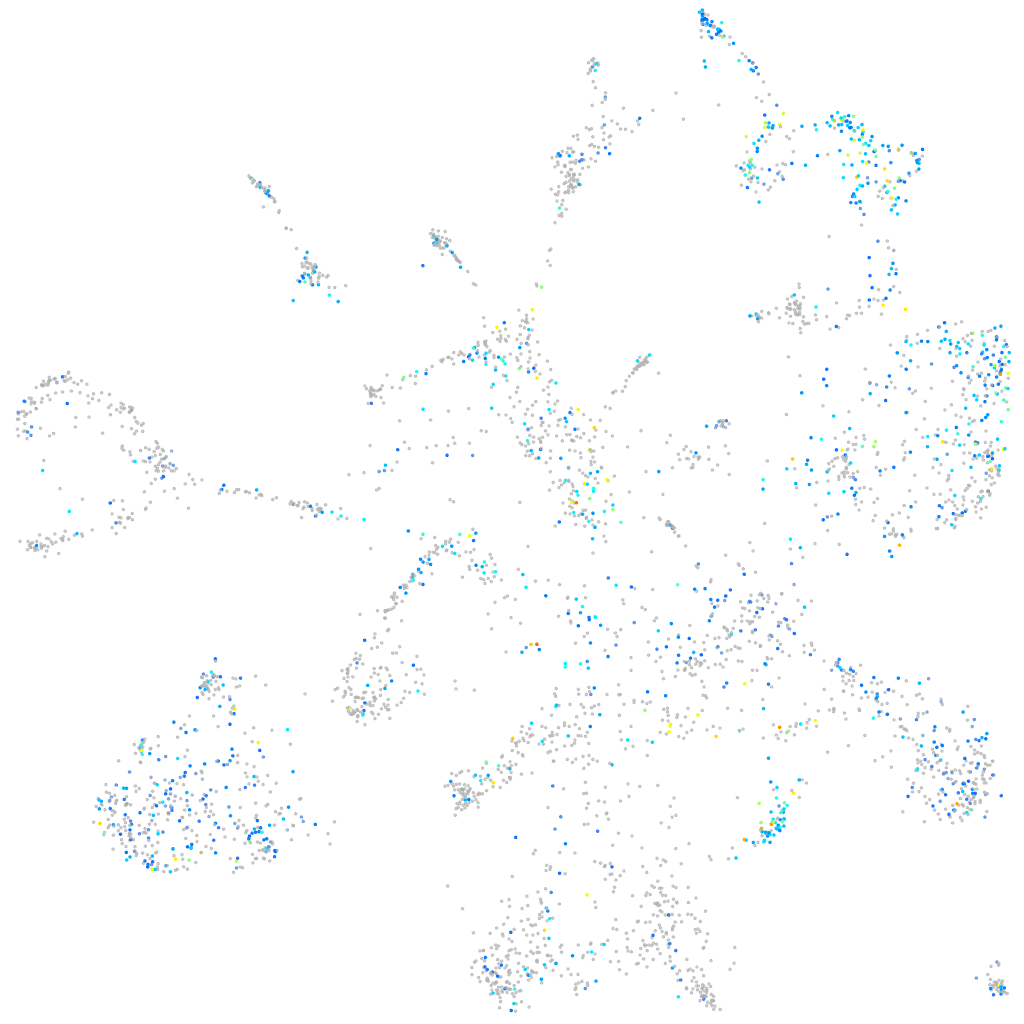
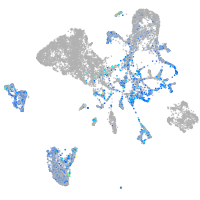
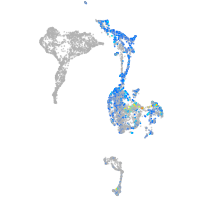
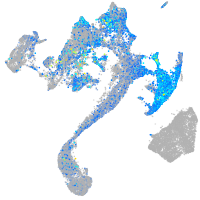
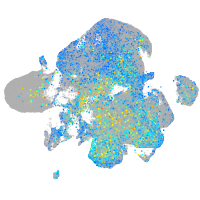
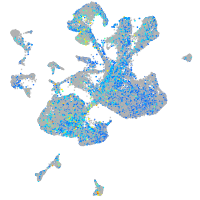
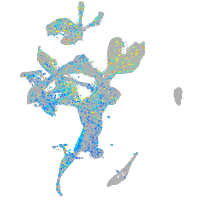
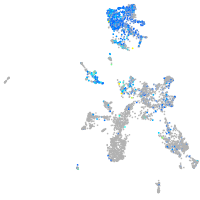
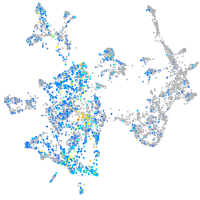
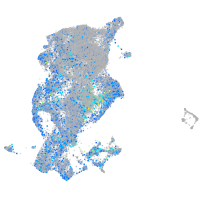
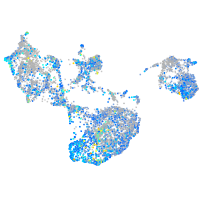
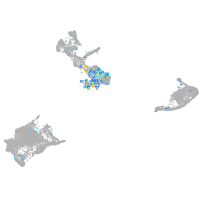
EPH receptor B3
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| alpi.1 | 0.250 | tpm1 | -0.195 |
| LO016987.2 | 0.232 | gapdhs | -0.190 |
| sfrp5 | 0.231 | myl9a | -0.186 |
| mmel1 | 0.231 | tagln | -0.186 |
| synpo2lb | 0.230 | acta2 | -0.184 |
| podxl | 0.214 | myl6 | -0.170 |
| lmod2a | 0.210 | myh11a | -0.165 |
| tmem88b | 0.207 | csrp1b | -0.161 |
| lbx2 | 0.204 | lmod1b | -0.161 |
| smoc1 | 0.199 | gpia | -0.157 |
| alx4a | 0.186 | eno3 | -0.149 |
| bnc2 | 0.185 | tpi1b | -0.148 |
| cxadr | 0.183 | tpm2 | -0.145 |
| jam2b | 0.183 | cald1b | -0.145 |
| krt94 | 0.178 | si:ch211-62a1.3 | -0.140 |
| eppk1 | 0.169 | cox6b1 | -0.137 |
| jcada | 0.167 | mylkb | -0.136 |
| COLEC10 | 0.166 | ppiab | -0.135 |
| adma | 0.161 | selenom | -0.134 |
| slc29a1b | 0.159 | ak1 | -0.134 |
| mdka | 0.157 | desmb | -0.133 |
| tnni1b | 0.156 | fbxl22 | -0.132 |
| evx1 | 0.156 | tuba8l2 | -0.131 |
| gata5 | 0.155 | cnn1b | -0.129 |
| smad9 | 0.154 | BX088707.3 | -0.126 |
| XLOC-042222 | 0.154 | ckba | -0.124 |
| tbx1 | 0.154 | smtna | -0.124 |
| crb2a | 0.152 | foxf1 | -0.123 |
| tbx18 | 0.151 | atp5mc1 | -0.123 |
| nr0b2a | 0.151 | pfn1 | -0.123 |
| bambia | 0.150 | acta1a | -0.123 |
| CABZ01075068.1 | 0.149 | calm3b | -0.122 |
| lrp2a | 0.148 | lpp | -0.122 |
| bcam | 0.148 | actb1 | -0.122 |
| krt8 | 0.148 | hoxc8a | -0.122 |