enolase superfamily member 1
ZFIN 
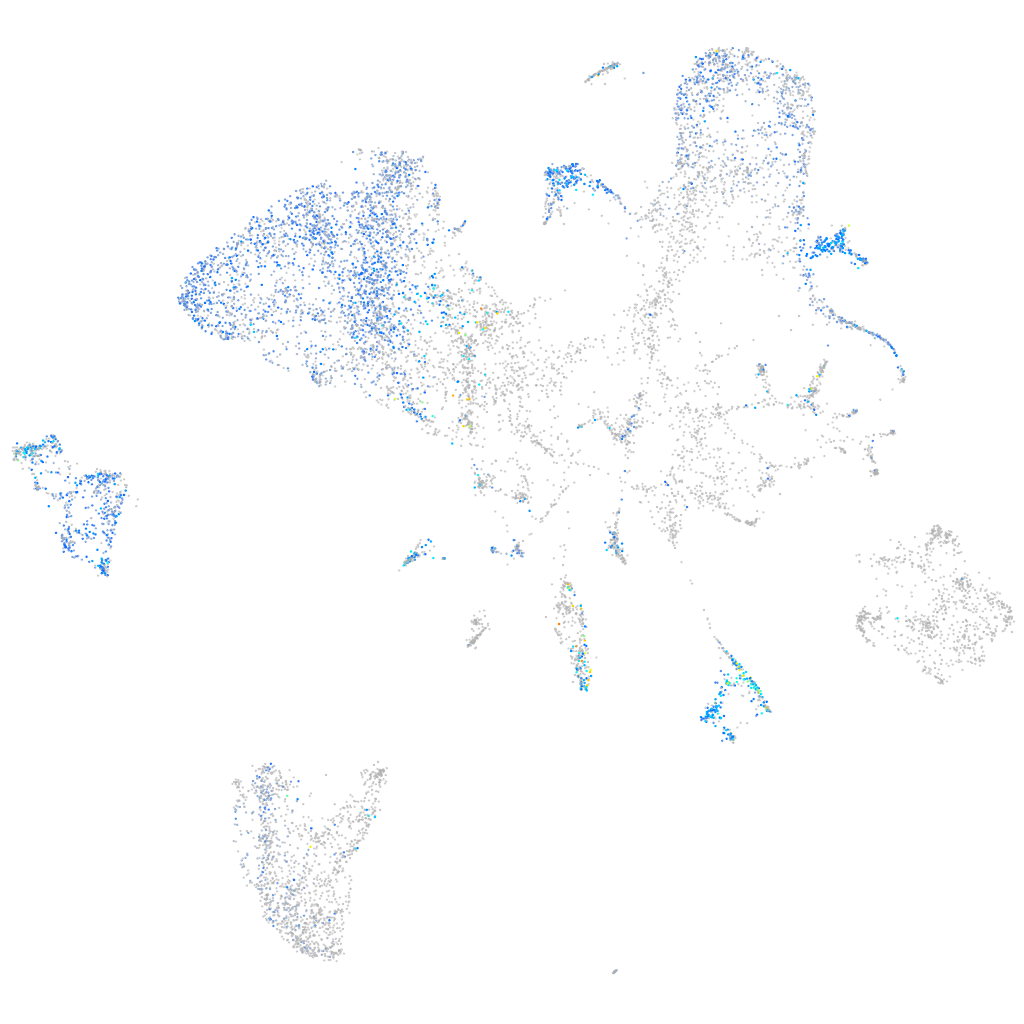
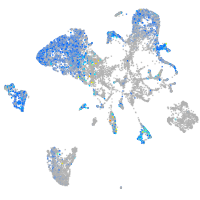
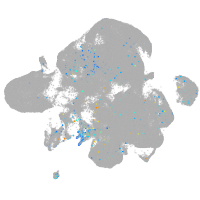
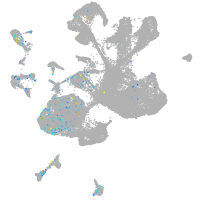
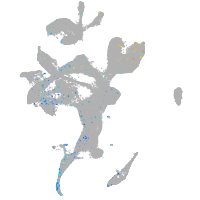
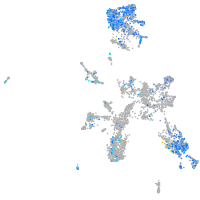
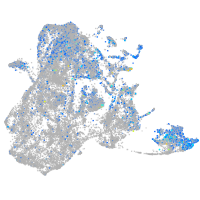
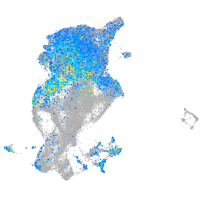
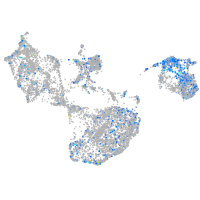
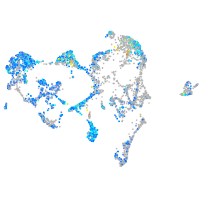
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| abca12 | 0.276 | npm1a | -0.204 |
| anxa3b | 0.270 | ncl | -0.203 |
| cabp2b | 0.265 | si:ch211-222l21.1 | -0.197 |
| sftpba | 0.257 | nop58 | -0.197 |
| anxa1a | 0.256 | dkc1 | -0.197 |
| ceacam1 | 0.255 | nucks1a | -0.188 |
| gstp1 | 0.254 | hmga1a | -0.184 |
| anxa5b | 0.253 | seta | -0.183 |
| lgals3b | 0.252 | prmt1 | -0.176 |
| txn | 0.251 | marcksb | -0.176 |
| b3gnt5a | 0.251 | abcf1 | -0.176 |
| si:dkey-262k9.2 | 0.249 | apex1 | -0.175 |
| itm2bb | 0.247 | fbl | -0.175 |
| npc2 | 0.245 | nop56 | -0.175 |
| dap1b | 0.244 | cirbpa | -0.172 |
| ahnak | 0.243 | hnrnpabb | -0.170 |
| s100v1 | 0.240 | snu13b | -0.169 |
| anxa11b | 0.240 | setb | -0.169 |
| glud1a | 0.240 | hmgb2b | -0.168 |
| agr2 | 0.238 | marcksl1b | -0.167 |
| s100a10b | 0.237 | snrpb | -0.166 |
| s100v2 | 0.236 | syncrip | -0.165 |
| cavin2b | 0.234 | sumo3b | -0.164 |
| capns1a | 0.233 | ddx18 | -0.164 |
| ctsz | 0.232 | nop2 | -0.164 |
| tent5bb | 0.231 | anp32b | -0.163 |
| cavin2a | 0.231 | cbx3a | -0.163 |
| gna15.1 | 0.231 | NC-002333.4 | -0.162 |
| pmp22b | 0.230 | khdrbs1a | -0.161 |
| BX908782.3 | 0.228 | hnrnpub | -0.160 |
| fn1b | 0.227 | hmgb1b | -0.159 |
| cebpd | 0.225 | hnrnpa0b | -0.158 |
| zgc:63831 | 0.225 | hnrnpaba | -0.158 |
| malb | 0.224 | cbx1a | -0.157 |
| krt91 | 0.221 | hnrnpa1b | -0.157 |