"enolase 3, (beta, muscle)"
ZFIN 
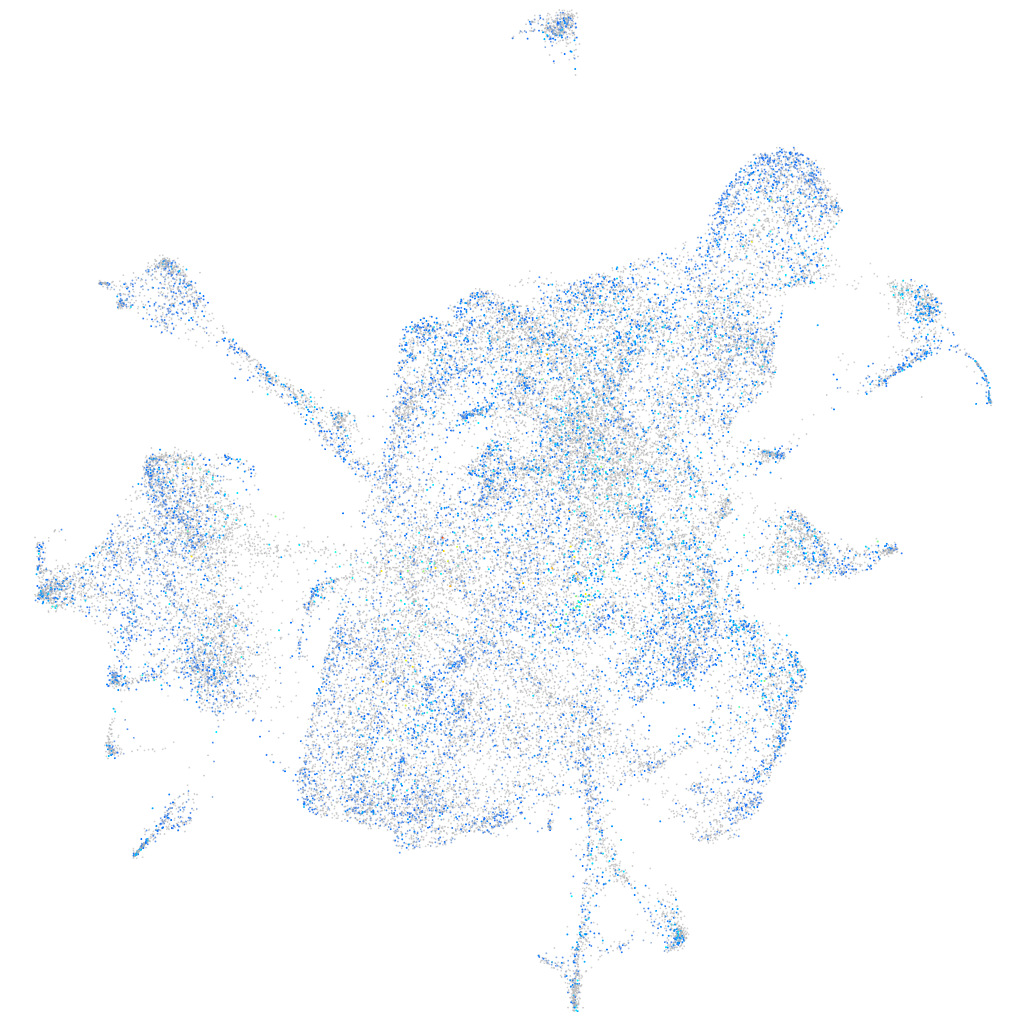
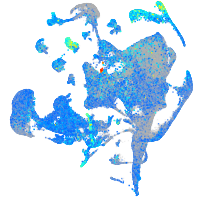
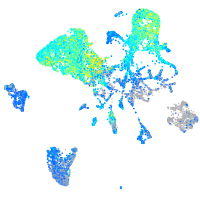
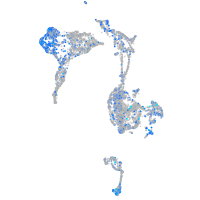
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:162944 | 0.098 | NC-002333.4 | -0.039 |
| zgc:153867 | 0.091 | cdh11 | -0.038 |
| GCA | 0.089 | CABZ01075068.1 | -0.037 |
| gapdhs | 0.087 | cd248b | -0.035 |
| arpc3 | 0.086 | NC-002333.17 | -0.033 |
| tpi1b | 0.085 | cxcl12a | -0.032 |
| atp5f1b | 0.085 | CABZ01072614.1 | -0.032 |
| dad1 | 0.084 | zbtb16a | -0.032 |
| ldhba | 0.084 | lin28a | -0.031 |
| atp5mc1 | 0.084 | ephb3 | -0.030 |
| sod1 | 0.083 | nr6a1a | -0.030 |
| nme2b.1 | 0.083 | col11a1b | -0.029 |
| pfn1 | 0.083 | mn1a | -0.028 |
| atp5fa1 | 0.082 | ednrab | -0.027 |
| si:ch211-156j16.1 | 0.081 | LOC110439372 | -0.026 |
| capzb | 0.080 | wu:fb97g03 | -0.026 |
| msna | 0.079 | BX927258.1 | -0.026 |
| pdia3 | 0.079 | XLOC-042222 | -0.025 |
| atp5pb | 0.078 | twist1a | -0.025 |
| xbp1 | 0.078 | ebf1a | -0.025 |
| arpc2 | 0.077 | mir199-2 | -0.025 |
| prdx2 | 0.077 | rps29 | -0.024 |
| gpia | 0.077 | elavl3 | -0.024 |
| hoxc8a | 0.076 | rpl38 | -0.024 |
| calm3b | 0.076 | XLOC-039121 | -0.023 |
| atp5pd | 0.076 | nectin1b | -0.023 |
| ostc | 0.076 | thbs2b | -0.023 |
| atp5mc3b | 0.075 | abi3bpa | -0.022 |
| vdac2 | 0.075 | npb | -0.022 |
| vdac3 | 0.075 | col5a1 | -0.022 |
| ppib | 0.075 | ebf3a | -0.022 |
| cst14a.2 | 0.075 | zgc:158463 | -0.021 |
| ywhabb | 0.074 | aspn | -0.021 |
| hoxb8a | 0.074 | col12a1a | -0.021 |
| capns1b | 0.074 | CR354435.1 | -0.021 |