EH-domain containing 1a
ZFIN 
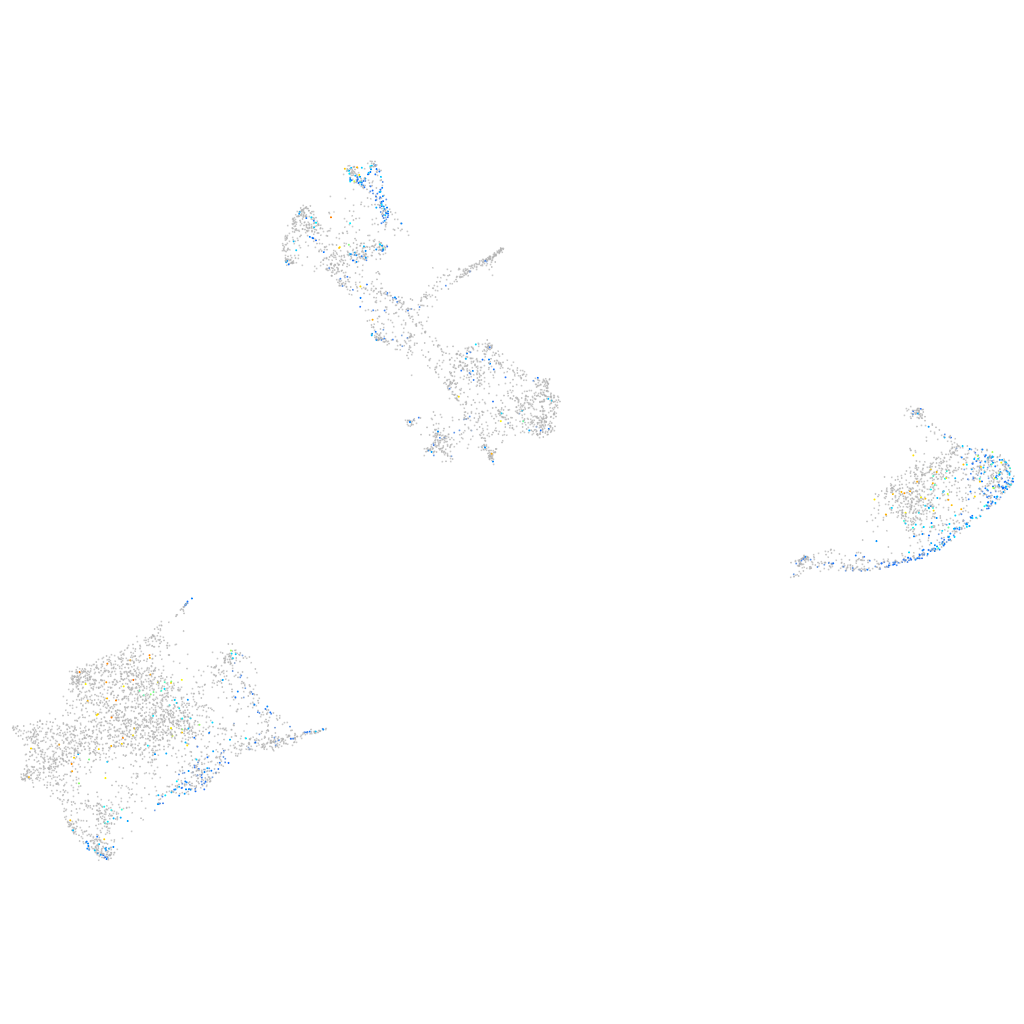
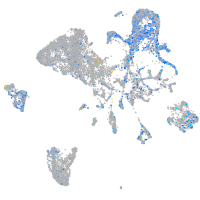
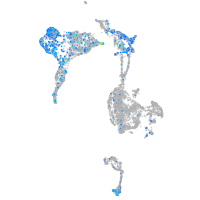
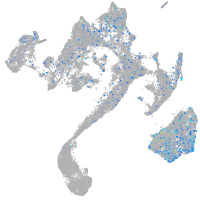
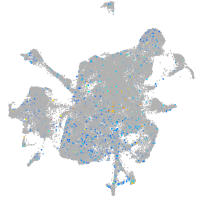
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.162 | CABZ01021592.1 | -0.097 |
| slc45a2 | 0.157 | si:ch211-222l21.1 | -0.074 |
| oca2 | 0.157 | marcksl1a | -0.072 |
| kita | 0.156 | si:ch73-1a9.3 | -0.071 |
| tyrp1a | 0.155 | ptmaa | -0.071 |
| dct | 0.154 | gpm6aa | -0.066 |
| tyrp1b | 0.150 | nova2 | -0.064 |
| pmela | 0.148 | tuba1c | -0.063 |
| slc39a10 | 0.148 | elavl3 | -0.062 |
| rab38 | 0.148 | hbae3 | -0.060 |
| yjefn3 | 0.147 | si:dkey-251i10.2 | -0.058 |
| atp6v0ca | 0.147 | epb41a | -0.055 |
| prkar1b | 0.145 | tmsb | -0.055 |
| mtbl | 0.144 | acbd7 | -0.055 |
| si:ch73-389b16.1 | 0.144 | tmeff1b | -0.053 |
| mlpha | 0.143 | h3f3a | -0.053 |
| slc29a3 | 0.140 | hmgn2 | -0.051 |
| slc37a2 | 0.140 | sox11b | -0.051 |
| slc7a5 | 0.140 | hmgb1b | -0.051 |
| slc24a5 | 0.139 | hbbe1.3 | -0.050 |
| si:ch211-195b13.1 | 0.139 | elavl4 | -0.049 |
| zgc:91968 | 0.138 | stmn1a | -0.049 |
| spra | 0.137 | pcna | -0.049 |
| lgals2a | 0.136 | cadm3 | -0.049 |
| aadac | 0.135 | atp1a3a | -0.049 |
| slc22a2 | 0.133 | zc4h2 | -0.048 |
| tspan36 | 0.133 | hnrnpaba | -0.048 |
| atp11a | 0.132 | cx43.4 | -0.048 |
| tyr | 0.130 | tmem130 | -0.048 |
| bace2 | 0.130 | hmgn3 | -0.048 |
| lamp1a | 0.129 | ttn.2 | -0.048 |
| slc3a2a | 0.128 | hmga1a | -0.048 |
| ift27 | 0.127 | vamp2 | -0.047 |
| tspan10 | 0.127 | CU467822.1 | -0.047 |
| anxa2a | 0.127 | apex1 | -0.047 |