ephrin-B3b
ZFIN 
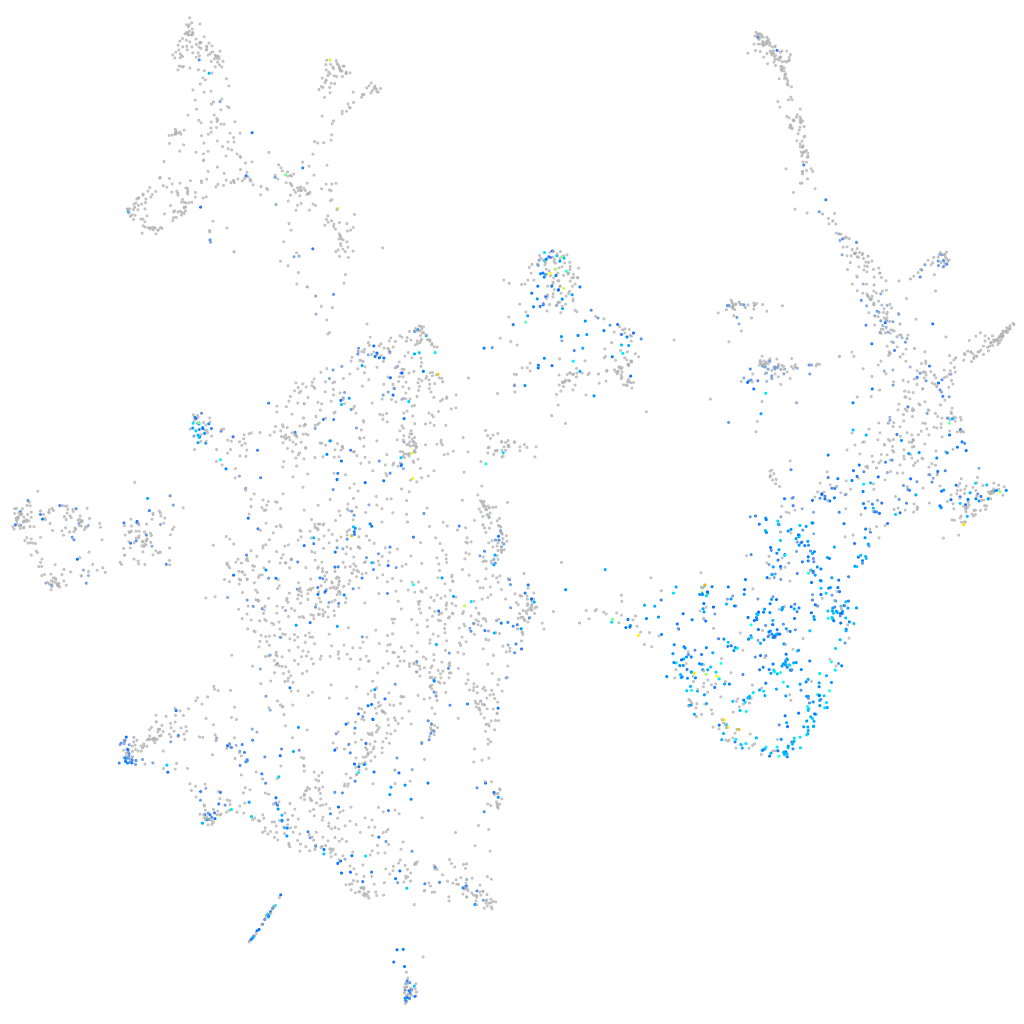
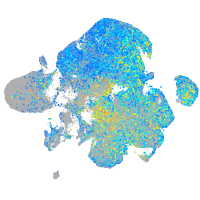
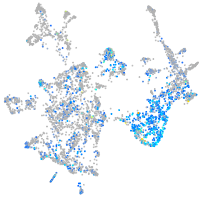
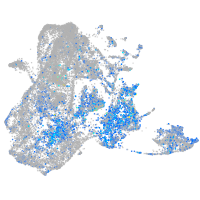
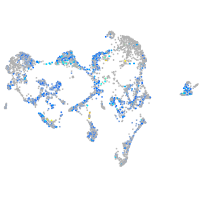
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tfap2a | 0.394 | si:ch211-243g18.2 | -0.300 |
| mcama | 0.386 | stm | -0.219 |
| pcdh12 | 0.344 | agr2 | -0.205 |
| sostdc1b | 0.341 | tspan13b | -0.196 |
| fat1b | 0.337 | eno1a | -0.195 |
| itga6b | 0.313 | gapdhs | -0.190 |
| cxcr4b | 0.304 | otofb | -0.181 |
| met | 0.302 | rbm24a | -0.178 |
| vangl1 | 0.296 | gpx1b | -0.177 |
| sfrp1a | 0.296 | gpx2 | -0.176 |
| ackr3b | 0.283 | atp1a3b | -0.174 |
| krt15 | 0.281 | CR925719.1 | -0.173 |
| plecb | 0.277 | osbpl1a | -0.173 |
| foxp4 | 0.276 | calml4a | -0.171 |
| marcksl1b | 0.275 | cd164l2 | -0.171 |
| celsr1b | 0.275 | tbx2a | -0.167 |
| lmo4b | 0.273 | atp2b1a | -0.166 |
| hhip | 0.273 | dnajc5b | -0.166 |
| col28a2a | 0.271 | otomp | -0.164 |
| mir205 | 0.269 | emb | -0.164 |
| tfap2c | 0.268 | apba1a | -0.163 |
| bzw2 | 0.265 | prr15la | -0.162 |
| inka1b | 0.265 | vsig8a | -0.161 |
| fxyd6l | 0.263 | si:ch73-15n15.3 | -0.161 |
| cyt1 | 0.263 | nptna | -0.161 |
| meis1a | 0.261 | spa17 | -0.161 |
| cyt1l | 0.260 | zgc:55461 | -0.161 |
| anxa4 | 0.254 | gipc3 | -0.161 |
| inka1a | 0.253 | daw1 | -0.161 |
| krt8 | 0.251 | pax2a | -0.160 |
| krt18b | 0.247 | bricd5 | -0.159 |
| si:ch211-202p1.5 | 0.246 | mb | -0.158 |
| she | 0.245 | pkma | -0.158 |
| robo1 | 0.244 | slc17a8 | -0.158 |
| plpp2b | 0.244 | lrrc73 | -0.157 |